5HAF
 
 | |
5IFR
 
 | | Structure of the stable UBE2D3-UbDha conjugate | | Descriptor: | GLYCEROL, Polyubiquitin-B, Ubiquitin-conjugating enzyme E2 D3 | | Authors: | Pruneda, J.N, Mulder, M.P.C, Witting, K, Ovaa, H, Komander, D. | | Deposit date: | 2016-02-26 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A cascading activity-based probe sequentially targets E1-E2-E3 ubiquitin enzymes.
Nat.Chem.Biol., 12, 2016
|
|
5HAG
 
 | |
9AZJ
 
 | |
9B0B
 
 | |
9B0Z
 
 | |
9AVW
 
 | |
9B12
 
 | |
9AVT
 
 | | Structure of TAB2 NZF domain bound to K6 / Lys6-linked diubiquitin | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, SULFATE ION, TGF-beta-activated kinase 1 and MAP3K7-binding protein 2, ... | | Authors: | Michel, M.A, Scutts, S, Komander, D. | | Deposit date: | 2024-03-04 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of TAB2 NZF domain bound to K6 / Lys6-linked diubiquitin
To be published
|
|
5HAM
 
 | |
5C9V
 
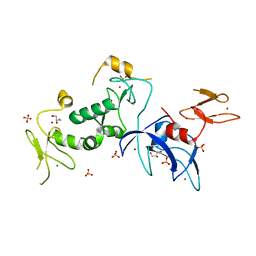 | | Structure of human Parkin G319A | | Descriptor: | E3 ubiquitin-protein ligase parkin, GLYCEROL, SULFATE ION, ... | | Authors: | Wauer, T, Komander, D. | | Deposit date: | 2015-06-29 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of phospho-ubiquitin-induced PARKIN activation.
Nature, 524, 2015
|
|
7TZJ
 
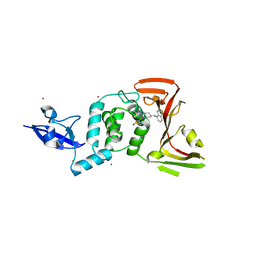 | | SARS CoV-2 PLpro in complex with inhibitor 3k | | Descriptor: | DIMETHYL SULFOXIDE, N-[(3-fluorophenyl)methyl]-1-[(1R)-1-naphthalen-1-ylethyl]piperidine-4-carboxamide, Papain-like protease, ... | | Authors: | Calleja, D.J, Klemm, T, Lechtenberg, B.C, Kuchel, N.W, Lessene, G, Komander, D. | | Deposit date: | 2022-02-15 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Insights Into Drug Repurposing, as Well as Specificity and Compound Properties of Piperidine-Based SARS-CoV-2 PLpro Inhibitors.
Front Chem, 10, 2022
|
|
7T4M
 
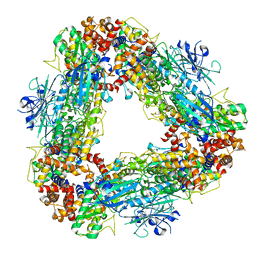 | | Structure of dodecameric unphosphorylated Pediculus humanus (Ph) PINK1 D357A mutant | | Descriptor: | Serine/threonine-protein kinase PINK1, putative | | Authors: | Gan, Z.Y, Leis, A, Dewson, G, Glukhova, A, Komander, D. | | Deposit date: | 2021-12-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Activation mechanism of PINK1.
Nature, 602, 2022
|
|
7T3X
 
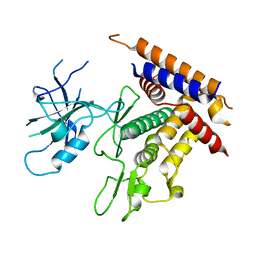 | | Structure of unphosphorylated Pediculus humanus (Ph) PINK1 D334A mutant | | Descriptor: | Serine/threonine-protein kinase PINK1 | | Authors: | Gan, Z.Y, Leis, A, Dewson, G, Glukhova, A, Komander, D. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | Activation mechanism of PINK1.
Nature, 602, 2022
|
|
7T4K
 
 | | Structure of dimeric phosphorylated Pediculus humanus (Ph) PINK1 with kinked alpha-C helix in chain B | | Descriptor: | Serine/threonine-protein kinase PINK1, putative | | Authors: | Gan, Z.Y, Leis, A, Dewson, G, Glukhova, A, Komander, D. | | Deposit date: | 2021-12-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Activation mechanism of PINK1.
Nature, 602, 2022
|
|
7T4N
 
 | | Structure of dimeric unphosphorylated Pediculus humanus (Ph) PINK1 D357A mutant | | Descriptor: | Serine/threonine-protein kinase PINK1, putative | | Authors: | Gan, Z.Y, Leis, A, Dewson, G, Glukhova, A, Komander, D. | | Deposit date: | 2021-12-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Activation mechanism of PINK1.
Nature, 602, 2022
|
|
7T4L
 
 | | Structure of dimeric phosphorylated Pediculus humanus (Ph) PINK1 with extended alpha-C helix in chain B | | Descriptor: | Serine/threonine-protein kinase PINK1, putative | | Authors: | Gan, Z.Y, Leis, A, Dewson, G, Glukhova, A, Komander, D. | | Deposit date: | 2021-12-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Activation mechanism of PINK1.
Nature, 602, 2022
|
|
9CYB
 
 | | SARS-CoV-2 PLpro in complex with inhibitor WEHI-P1 | | Descriptor: | Papain-like protease, SUCCINIC ACID, [(3R)-1-cyclopentylpiperidin-3-yl](6-methoxynaphthalen-2-yl)methanone | | Authors: | Calleja, D.J, Lechtenberg, B.C, Kuchel, N.W, Devine, S.M, Bader, S.M, Doerflinger, M, Mitchell, J.P, Lessene, G, Komander, D. | | Deposit date: | 2024-08-01 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A novel PLpro inhibitor improves outcomes in a pre-clinical model of long COVID.
Nat Commun, 16, 2025
|
|
9CYD
 
 | | SARS-CoV-2 PLpro in complex with inhibitor WEHI-P4 | | Descriptor: | (1S,4s)-4-{(3R)-3-[(E)-(methoxyimino)(6-methoxynaphthalen-2-yl)methyl]piperidin-1-yl}cyclohexan-1-ol, CHLORIDE ION, Papain-like protease, ... | | Authors: | Calleja, D.J, Lechtenberg, B.C, Kuchel, N.W, Devine, S.M, Bader, S.M, Doerflinger, M, Mitchell, J.P, Lessene, G, Komander, D. | | Deposit date: | 2024-08-02 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A novel PLpro inhibitor improves outcomes in a pre-clinical model of long COVID.
Nat Commun, 16, 2025
|
|
9CYK
 
 | | SARS-CoV-2 PLpro in complex with inhibitor WEHI-P24 | | Descriptor: | ACETIC ACID, GLYCEROL, Papain-like protease, ... | | Authors: | Calleja, D.J, Lechtenberg, B.C, Kuchel, N.W, Devine, S.M, Bader, S.M, Doerflinger, M, Mitchell, J.P, Lessene, G, Komander, D. | | Deposit date: | 2024-08-02 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A novel PLpro inhibitor improves outcomes in a pre-clinical model of long COVID.
Nat Commun, 16, 2025
|
|
9CYC
 
 | | SARS-CoV-2 PLpro in complex with inhibitor WEHI-P2 | | Descriptor: | (E)-1-[(3R)-1-cyclopentylpiperidin-3-yl]-N-methoxy-1-(6-methoxynaphthalen-2-yl)methanimine, ACETIC ACID, GLYCEROL, ... | | Authors: | Calleja, D.J, Lechtenberg, B.C, Kuchel, N.W, Devine, S.M, Bader, S.M, Doerflinger, M, Mitchel, J.P, Lessene, G, Komander, D. | | Deposit date: | 2024-08-01 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A novel PLpro inhibitor improves outcomes in a pre-clinical model of long COVID.
Nat Commun, 16, 2025
|
|
6FFA
 
 | | FMDV Leader protease bound to substrate ISG15 | | Descriptor: | GLYCEROL, Lbpro, SULFATE ION, ... | | Authors: | Swatek, K.N, Pruneda, J.N, Komander, D. | | Deposit date: | 2018-01-05 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Irreversible inactivation of ISG15 by a viral leader protease enables alternative infection detection strategies.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6GZT
 
 | |
6GLC
 
 | | Structure of phospho-Parkin bound to phospho-ubiquitin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | Gladkova, C, Maslen, S.L, Skehel, J.M, Komander, D. | | Deposit date: | 2018-05-23 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of parkin activation by PINK1.
Nature, 559, 2018
|
|
6GZU
 
 | |
