2N8F
 
 | |
2NDO
 
 | | Structure of EcDsbA-sulfonamide1 complex | | Descriptor: | 2-{[(4-iodophenyl)sulfonyl]amino}benzoic acid, Thiol:disulfide interchange protein DsbA | | Authors: | Williams, M.L, Doak, B.C, Vazirani, M, Ilyichova, O, Wang, G, Bermel, W, Simpson, J.S, Chalmers, D.K, King, G.F, Mobli, M, Scanlon, M.J. | | Deposit date: | 2016-08-22 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Determination of ligand binding modes in weak protein-ligand complexes using sparse NMR data.
J.Biomol.Nmr, 66, 2016
|
|
2MFA
 
 | | Mambalgin-2 | | Descriptor: | Mambalgin-2 | | Authors: | Schroeder, C.I, Rash, L.D, Vila-Farres, X, Rosengren, K.J, Mobli, M, King, G.F, Alewood, P.F, Craik, D.J, Durek, T. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Chemical synthesis, 3D structure, and ASIC binding site of the toxin mambalgin-2.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6OFA
 
 | | Wasabi Receptor Toxin | | Descriptor: | Wasabi Receptor Toxin | | Authors: | Lin King, J.V, Kelly, M.J.S, Julius, D. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | A Cell-Penetrating Scorpion Toxin Enables Mode-Specific Modulation of TRPA1 and Pain.
Cell, 178, 2019
|
|
4ZIJ
 
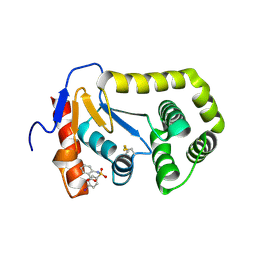 | | Crystal structure of E.Coli DsbA in complex with 2-(4-iodophenylsulfonamido) benzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(4-iodophenyl)sulfonyl]amino}benzoic acid, Thiol:disulfide interchange protein DsbA | | Authors: | Vazirani, M, Ilyichova, O.V, Scanlon, M.J. | | Deposit date: | 2015-04-28 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Determination of ligand binding modes in weak protein-ligand complexes using sparse NMR data.
J.Biomol.Nmr, 66, 2016
|
|
6OVJ
 
 | |
6PX7
 
 | |
6PX8
 
 | |
6OTA
 
 | |
6OTB
 
 | |
3LUI
 
 | | Crystal structure of the SNX17 PX domain with bound sulphate | | Descriptor: | SULFATE ION, Sorting nexin-17 | | Authors: | Ghai, R, Collins, B.M. | | Deposit date: | 2010-02-17 | | Release date: | 2010-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Phox homology band 4.1/ezrin/radixin/moesin-like proteins function as molecular scaffolds that interact with cargo receptors and Ras GTPases
Proc.Natl.Acad.Sci.USA, 2011
|
|
6MY1
 
 | | Solution structure of gomesin at 278 K | | Descriptor: | gomesin | | Authors: | Chin, Y.K.-Y, Deplazes, E. | | Deposit date: | 2018-10-31 | | Release date: | 2019-11-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The unusual conformation of cross-strand disulfide bonds is critical to the stability of beta-hairpin peptides.
Proteins, 88, 2020
|
|
6MY2
 
 | | Solution structure of gomesin at 298 K | | Descriptor: | gomesin | | Authors: | Chin, Y.K.-Y, Deplazes, E. | | Deposit date: | 2018-10-31 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The unusual conformation of cross-strand disulfide bonds is critical to the stability of beta-hairpin peptides.
Proteins, 88, 2020
|
|
6MY3
 
 | | Solution structure of gomesin at 310K | | Descriptor: | gomesin | | Authors: | Chin, Y.K.-Y, Deplazes, E. | | Deposit date: | 2018-11-01 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | The unusual conformation of cross-strand disulfide bonds is critical to the stability of beta-hairpin peptides.
Proteins, 88, 2020
|
|
5T3M
 
 | |
5T4R
 
 | |
5FZW
 
 | |
5FZX
 
 | |
5FZV
 
 | |
5ZFO
 
 | | NMR structure of IRD12 from Capsicum annum. | | Descriptor: | Pin-II type proteinase inhibitor 38 | | Authors: | Gartia, J, Barnwal, R.P, Chary, K.V.R. | | Deposit date: | 2018-03-06 | | Release date: | 2019-05-15 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NMR structure and dynamics of inhibitory repeat domain variant 12, a plant protease inhibitor from Capsicum annuum, and its structural relationship to other plant protease inhibitors.
J.Biomol.Struct.Dyn., 2019
|
|
7JPM
 
 | |
3OF6
 
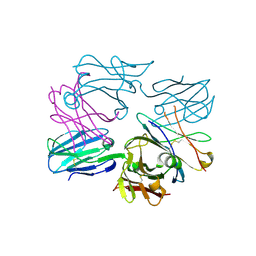 | | Human pre-T cell receptor crystal structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pre T-cell antigen receptor alpha, T cell receptor beta chain | | Authors: | Pang, S.S. | | Deposit date: | 2010-08-13 | | Release date: | 2010-10-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis for autonomous dimerization of the pre-T-cell antigen receptor
Nature, 467, 2010
|
|
7R6P
 
 | |
6BTV
 
 | | Solution NMR structures for CcoTx-II | | Descriptor: | Beta-theraphotoxin-Cm1b | | Authors: | Agwa, A.J, Schroeder, C.I. | | Deposit date: | 2017-12-07 | | Release date: | 2018-05-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Gating modifier toxins isolated from spider venom: Modulation of voltage-gated sodium channels and the role of lipid membranes.
J. Biol. Chem., 293, 2018
|
|
6BR0
 
 | | Solution NMR structure for CcoTx-I | | Descriptor: | Beta-theraphotoxin-Cm1a | | Authors: | Agwa, A.J, Schroeder, C.I. | | Deposit date: | 2017-11-29 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Gating modifier toxins isolated from spider venom: Modulation of voltage-gated sodium channels and the role of lipid membranes.
J. Biol. Chem., 293, 2018
|
|
