3D00
 
 | |
3KK7
 
 | |
3CGH
 
 | |
3CM1
 
 | |
2OOC
 
 | |
3EQX
 
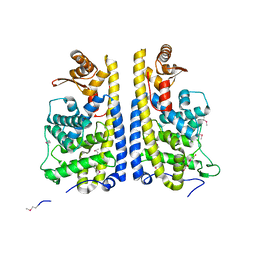 | |
2Q8U
 
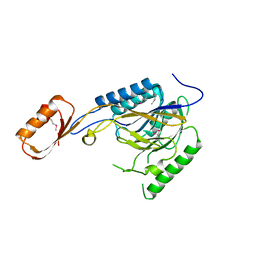 | |
2AYK
 
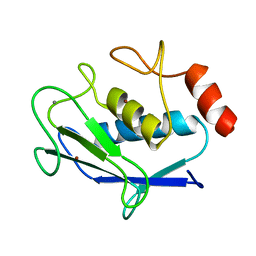 | |
4LY9
 
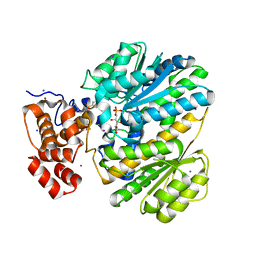 | | Human GKRP complexed to AMG-1694 [(2R)-1,1,1-trifluoro-2-{4-[(2S)-2-{[(3S)-3-methylmorpholin-4-yl]methyl}-4-(thiophen-2-ylsulfonyl)piperazin-1-yl]phenyl}propan-2-ol] and sorbitol-6-phosphate | | Descriptor: | (2R)-1,1,1-trifluoro-2-{4-[(2S)-2-{[(3S)-3-methylmorpholin-4-yl]methyl}-4-(thiophen-2-ylsulfonyl)piperazin-1-yl]phenyl}propan-2-ol, D-SORBITOL-6-PHOSPHATE, GLYCEROL, ... | | Authors: | Jordan, S.R. | | Deposit date: | 2013-07-30 | | Release date: | 2013-11-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Antidiabetic effects of glucokinase regulatory protein small-molecule disruptors.
Nature, 504, 2013
|
|
3H50
 
 | |
2QTP
 
 | |
3F1Z
 
 | |
3NL9
 
 | |
3DEE
 
 | |
3GF8
 
 | |
3H0N
 
 | |
2HBW
 
 | |
2RE3
 
 | |
3HSA
 
 | |
3L5O
 
 | |
4OHP
 
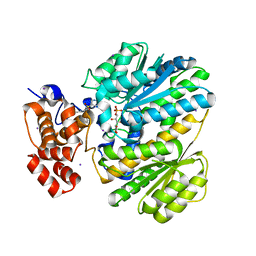 | | Human GKRP bound to AMG-3227 and S6P | | Descriptor: | 4-[(2S)-4-[(6-aminopyridin-3-yl)sulfonyl]-2-(prop-1-yn-1-yl)piperazin-1-yl]-N-methylbenzenesulfonamide, D-SORBITOL-6-PHOSPHATE, GLYCEROL, ... | | Authors: | Jordan, S.R, Chmait, S. | | Deposit date: | 2014-01-17 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Small molecule disruptors of the glucokinase-glucokinase regulatory protein interaction: 3. Structure-activity relationships within the aryl carbinol region of the N-arylsulfonamido-N'-arylpiperazine series.
J.Med.Chem., 57, 2014
|
|
4OHM
 
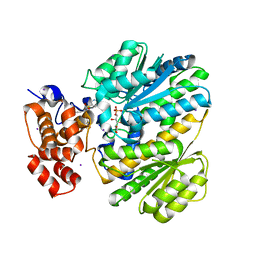 | | Human GKRP bound to AMG-0771 and sorbitol-6-phosphate | | Descriptor: | (2S)-2-{4-[(2S)-4-[(6-aminopyridin-3-yl)sulfonyl]-2-(prop-1-yn-1-yl)piperazin-1-yl]phenyl}-3,3,3-trifluoropropane-1,2-diol, D-SORBITOL-6-PHOSPHATE, GLYCEROL, ... | | Authors: | Jordan, S.R, Chmait, S. | | Deposit date: | 2014-01-17 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Small molecule disruptors of the glucokinase-glucokinase regulatory protein interaction: 3. Structure-activity relationships within the aryl carbinol region of the N-arylsulfonamido-N'-arylpiperazine series.
J.Med.Chem., 57, 2014
|
|
4OHO
 
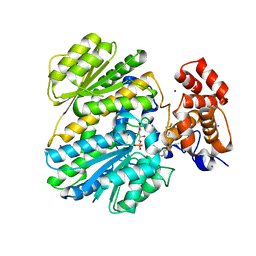 | | Human GKRP bound to AMG-2668 | | Descriptor: | 5-{[(3S)-3-(prop-1-yn-1-yl)-4-{4-[S-(trifluoromethyl)sulfonimidoyl]phenyl}piperazin-1-yl]sulfonyl}pyridin-2-amine, D-SORBITOL-6-PHOSPHATE, GLYCEROL, ... | | Authors: | Jordan, S.R, Chmait, S. | | Deposit date: | 2014-01-17 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Small molecule disruptors of the glucokinase-glucokinase regulatory protein interaction: 3. Structure-activity relationships within the aryl carbinol region of the N-arylsulfonamido-N'-arylpiperazine series.
J.Med.Chem., 57, 2014
|
|
1AYK
 
 | |
2IIZ
 
 | |
