8H8H
 
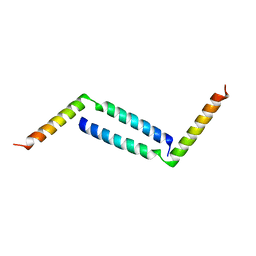 | |
5FTQ
 
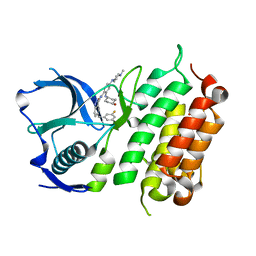 | | Crystal structure of the ALK kinase domain in complex with Cmpd 17 | | Descriptor: | ALK TYROSINE KINASE RECEPTOR, GLYCEROL, N-[5-(3,5-DIFLUOROBENZYL)-1H-INDAZOL-3-YL]-2-[(4-HYDROXYCYCLOHEXYL)AMINO]-4-(4-METHYLPIPERAZIN-1-YL) BENZAMIDE | | Authors: | Bossi, R, Canevari, G, Fasolini, M, Menichincheri, M, Ardini, E, Magnaghi, P, Avanzi, N, Banfi, P, Buffa, L, Ceriani, L, Colombo, M, Corti, L, Donati, D, Felder, E, Fiorelli, C, Fiorentini, F, Galvani, A, Isacchi, A, Lombardi Borgia, A, Marchionni, C, Nesi, M, Orrenius, C, Panzeri, A, Perrone, E, Pesenti, E, Rusconi, L, Saccardo, M.B, Vanotti, E, Orsini, P. | | Deposit date: | 2016-01-14 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Entrectinib: A New 3-Aminoindazole as a Potent Anaplastic Lymphoma Kinase (Alk), C-Ros Oncogene 1 Kinase (Ros1), and Pan-Tropomyosin Receptor Kinases (Pan-Trks) Inhibitor.
J.Med.Chem., 59, 2016
|
|
5FTO
 
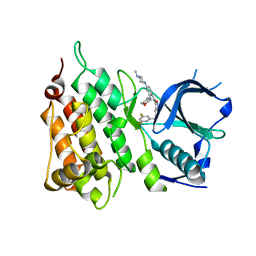 | | Crystal structure of the ALK kinase domain in complex with Entrectinib | | Descriptor: | ALK TYROSINE KINASE RECEPTOR, Entrectinib | | Authors: | Bossi, R, Canevari, G, Fasolini, M, Menichincheri, M, Ardini, E, Magnaghi, P, Avanzi, N, Banfi, P, Buffa, L, Ceriani, L, Colombo, M, Corti, L, Donati, D, Felder, E, Fiorelli, C, Fiorentini, F, Galvani, A, Isacchi, A, Lombardi Borgia, A, Marchionni, C, Nesi, M, Orrenius, C, Panzeri, A, Perrone, E, Pesenti, E, Rusconi, L, Saccardo, M.B, Vanotti, E, Orsini, P. | | Deposit date: | 2016-01-14 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery of Entrectinib: A New 3-Aminoindazole as a Potent Anaplastic Lymphoma Kinase (Alk), C-Ros Oncogene 1 Kinase (Ros1), and Pan-Tropomyosin Receptor Kinases (Pan-Trks) Inhibitor.
J.Med.Chem., 59, 2016
|
|
1NOE
 
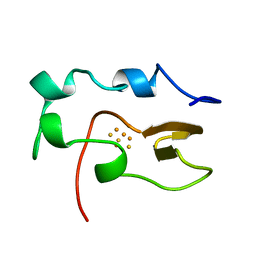 | | NMR STUDY OF REDUCED HIGH POTENTIAL IRON SULFUR PROTEIN | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Bentrop, D, Bertini, I, Capozzi, F, Dikiy, A, Eltis, L, Luchinat, C. | | Deposit date: | 1996-01-07 | | Release date: | 1996-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the reduced C77S mutant of the Chromatium vinosum high-potential iron-sulfur protein through nuclear magnetic resonance: comparison with the solution structure of the wild-type protein.
Biochemistry, 35, 1996
|
|
4GUY
 
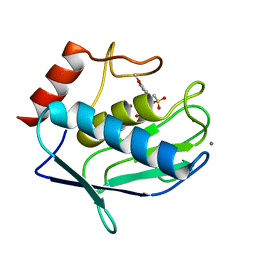 | | Human MMP12 catalytic domain in complex with*N*-Hydroxy-2-(2-(4-methoxyphenyl)ethylsulfonamido)acetamide | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, N-hydroxy-N~2~-{[2-(4-methoxyphenyl)ethyl]sulfonyl}glycinamide, ... | | Authors: | Calderone, V, Fragai, M, Luchinat, C, Massaro, A, Mordini, A, Mori, M. | | Deposit date: | 2012-08-30 | | Release date: | 2012-09-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contribution of free energy of solvation to ligand affinity in new potent MMPs inhibitors.
To be Published
|
|
1KSM
 
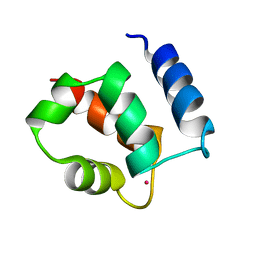 | | AVERAGE NMR SOLUTION STRUCTURE OF CA LN CALBINDIN D9K | | Descriptor: | LANTHANUM (III) ION, VITAMIN D-DEPENDENT CALCIUM-BINDING PROTEIN | | Authors: | Bertini, I, Donaire, A, Luchinat, C, Piccioli, M, Poggi, L, Parigi, G, Jimenez, B. | | Deposit date: | 2002-01-14 | | Release date: | 2002-01-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Paramagnetism-based versus classical constraints: an analysis of the solution structure of Ca Ln calbindin D9k.
J.Biomol.NMR, 21, 2001
|
|
5AE6
 
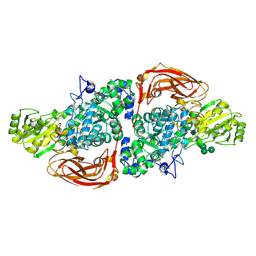 | | The structure of Hypocrea jecorina beta-xylosidase Xyl3A (Bxl1) in complex with 4-thioxylobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-XYLOSIDASE, ... | | Authors: | Mikkelsen, N.E, Gudmundsson, M, Karkehabadi, S, Hansson, H, Sandgren, M, Larenas, E, Mitchinson, C, Keleman, B, Kaper, T. | | Deposit date: | 2015-08-26 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Th Crystal Structure of a Fungal Glycoside Hydrolase Family 3 Beta-Xylosidase, Xyl3A from Hypocrea Jecorina
To be Published
|
|
4J2C
 
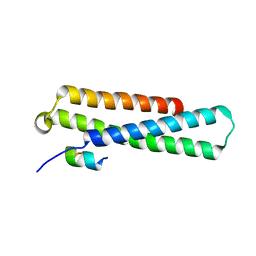 | | GARP-SNARE Interaction | | Descriptor: | Syntaxin-6, Vacuolar protein sorting-associated protein 51 homolog | | Authors: | Abascal-Palacios, G, Schindler, C, Rojas, A.L, Bonifacino, J.S, Hierro, A. | | Deposit date: | 2013-02-04 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis for the interaction of the Golgi-Associated Retrograde Protein Complex with the t-SNARE Syntaxin 6.
Structure, 21, 2013
|
|
5A7M
 
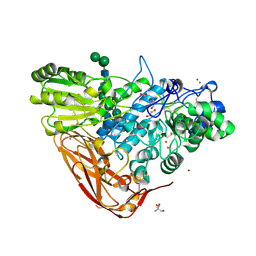 | | The structure of Hypocrea jecorina beta-xylosidase Xyl3A (Bxl1) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mikkelsen, N.E, Gudmundsson, M, Karkehabadi, S, Hansson, H, Sandgren, M, Larenas, E, Mitchinson, C, Keleman, B, Kaper, T. | | Deposit date: | 2015-07-08 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Th Crystal Structure of a Fungal Glycoside Hydrolase Family 3 Beta-Xylosidase, Xyl3A from Hypocrea Jecorina
To be Published
|
|
5FOJ
 
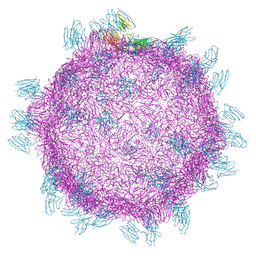 | | Cryo electron microscopy structure of Grapevine Fanleaf Virus complex with Nanobody | | Descriptor: | Nanobody, RNA2 polyprotein | | Authors: | Orlov, I, Hemmer, C, Ackerer, L, Lorber, B, Ghannam, A, Poignavent, V, Hleibieh, K, Sauter, C, Schmitt-Keichinger, C, Belval, L, Hily, J.M, Marmonier, A, Komar, V, Gersch, S, Schellenberger, P, Bron, P, Vigne, E, Muyldermans, S, Lemaire, O, Demangeat, G, Ritzenthaler, C, Klaholz, B.P. | | Deposit date: | 2015-11-22 | | Release date: | 2016-01-20 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of nanobody recognition of grapevine fanleaf virus and of virus resistance loss.
Proc.Natl.Acad.Sci.USA, 2020
|
|
1K0T
 
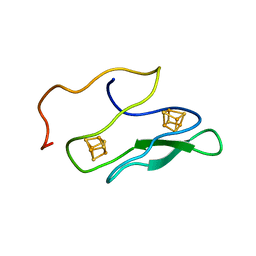 | | NMR SOLUTION STRUCTURE OF UNBOUND, OXIDIZED PHOTOSYSTEM I SUBUNIT PSAC, CONTAINING [4FE-4S] CLUSTERS FA AND FB | | Descriptor: | IRON/SULFUR CLUSTER, PSAC SUBUNIT OF PHOTOSYSTEM I | | Authors: | Antonkine, M.L, Liu, G, Bentrop, D, Bryant, D.A, Bertini, I, Luchinat, C, Golbeck, J.H, Stehlik, D. | | Deposit date: | 2001-09-20 | | Release date: | 2002-06-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the unbound, oxidized Photosystem I subunit PsaC, containing [4Fe-4S] clusters F(A) and F(B): a conformational change occurs upon binding to photosystem I.
J.Biol.Inorg.Chem., 7, 2002
|
|
1CLF
 
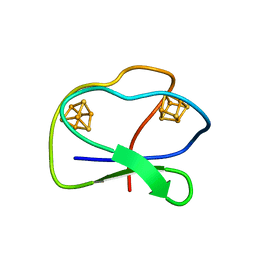 | | CLOSTRIDIUM PASTEURIANUM FERREDOXIN | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Bertini, I, Donaire, A, Feinberg, B.A, Luchinat, C, Piccioli, M, Yuan, H. | | Deposit date: | 1995-06-21 | | Release date: | 1996-01-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the oxidized 2[4Fe-4S] ferredoxin from Clostridium pasteurianum.
Eur.J.Biochem., 232, 1995
|
|
1KQV
 
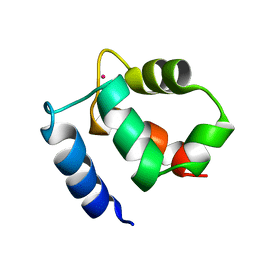 | | Family of NMR Solution Structures of Ca Ln Calbindin D9K | | Descriptor: | LANTHANUM (III) ION, VITAMIN D-DEPENDENT CALCIUM-BINDING PROTEIN | | Authors: | Bertini, I, Donaire, A, Jimenez, B, Luchinat, C, Parigi, G, Piccioli, M, Poggi, L. | | Deposit date: | 2002-01-08 | | Release date: | 2002-01-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Paramagnetism-based versus classical constraints: an analysis of the solution structure of Ca Ln calbindin D9k.
J.Biomol.NMR, 21, 2001
|
|
1HNS
 
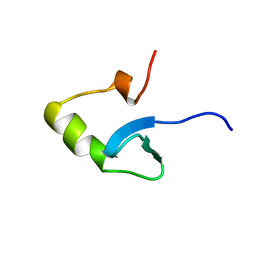 | | H-NS (DNA-BINDING DOMAIN) | | Descriptor: | H-NS | | Authors: | Shindo, H, Iwaki, T, Ieda, R, Kurumizaka, H, Ueguchi, C, Mizuno, T, Morikawa, S, Nakamura, H, Kuboniwa, H. | | Deposit date: | 1995-04-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from Escherichia coli.
FEBS Lett., 360, 1995
|
|
6ES4
 
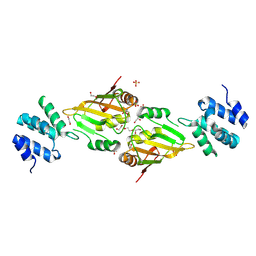 | | A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Syncrip, ... | | Authors: | Hobor, F, Dallmann, A, Ball, N.J, Cicchini, C, Battistelli, C, Ogrodowicz, R.W, Christodoulou, E, Martin, S.R, Castello, A, Tripodi, M, Taylor, I.A, Ramos, A. | | Deposit date: | 2017-10-19 | | Release date: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets.
Nat Commun, 9, 2018
|
|
1J5C
 
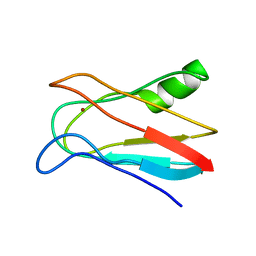 | | SOLUTION STRUCTURE OF OXIDIZED PARAMAGNETIC CU(II) PLASTOCYANIN FROM SYNECHOCYSTIS PCC6803 | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Bertini, I, Ciurli, S, Dikiy, A, Fernandez, C.O, Luchinat, C, Safarov, N, Shumilin, S, Vila, A.J. | | Deposit date: | 2002-04-02 | | Release date: | 2002-04-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The first solution structure of a paramagnetic copper(II) protein: the case of oxidized plastocyanin from the cyanobacterium Synechocystis PCC6803.
J.Am.Chem.Soc., 123, 2001
|
|
3CAR
 
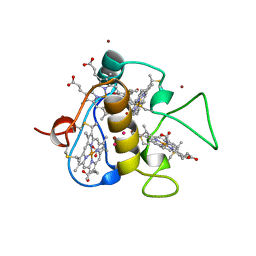 | | REDUCED STRUCTURE OF THE ACIDIC CYTOCHROME C3 FROM DESULFOVIBRIO AFRICANUS | | Descriptor: | ARSENIC, CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Norager, S, Legrand, P, Pieulle, L, Hatchikian, C, Roth, M. | | Deposit date: | 1998-11-17 | | Release date: | 2000-07-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the oxidised and reduced acidic cytochrome c3from Desulfovibrio africanus.
J.Mol.Biol., 290, 1999
|
|
3CAO
 
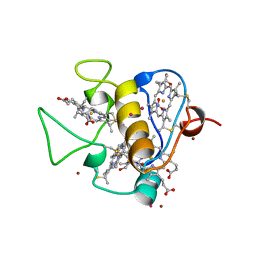 | | OXIDISED STRUCTURE OF THE ACIDIC CYTOCHROME C3 FROM DESULFOVIBRIO AFRICANUS | | Descriptor: | ARSENIC, CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Norager, S, Legrand, P, Pieulle, L, Hatchikian, C, Roth, M. | | Deposit date: | 1998-11-17 | | Release date: | 2000-07-23 | | Last modified: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the oxidised and reduced acidic cytochrome c3from Desulfovibrio africanus.
J.Mol.Biol., 290, 1999
|
|
1RJU
 
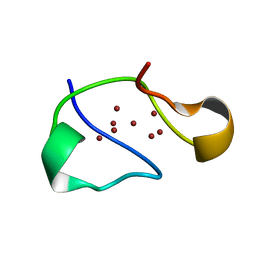 | | Crystal structure of a truncated form of yeast copper thionein | | Descriptor: | COPPER (I) ION, Metallothionein | | Authors: | Calderone, V, Dolderer, B, Hartmann, H.J, Echner, H, Luchinat, C, Del Bianco, C, Mangani, S, Weser, U. | | Deposit date: | 2003-11-20 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | The Crystal Structure of Yeast Copper Thionein: the solution of a long lasting Enigma.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1NEH
 
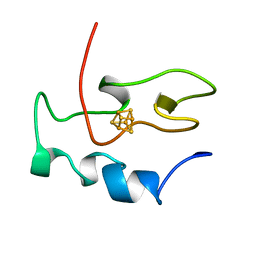 | | HIGH POTENTIAL IRON-SULFUR PROTEIN | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Bertini, I, Dikiy, A, Kastrau, D.H.W, Luchinat, C, Sompornpisut, P. | | Deposit date: | 1995-12-14 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the oxidized high potential iron-sulfur protein from Chromatium vinosum through NMR. Comparative analysis with the solution structure of the reduced species.
Biochemistry, 34, 1995
|
|
1GX7
 
 | | Best model of the electron transfer complex between cytochrome c3 and [Fe]-hydrogenase | | Descriptor: | 1,3-PROPANEDITHIOL, CARBON MONOXIDE, CYANIDE ION, ... | | Authors: | Elantak, L, Morelli, X, Bornet, O, Hatchikian, C, Czjzek, M, Dolla, A, Guerlesquin, F. | | Deposit date: | 2002-03-28 | | Release date: | 2003-07-31 | | Last modified: | 2019-11-27 | | Method: | SOLUTION NMR, THEORETICAL MODEL | | Cite: | The Cytochrome C(3)-[Fe]-Hydrogenase Electron-Transfer Complex: Structural Model by NMR Restrained Docking
FEBS Lett., 548, 2003
|
|
6SCZ
 
 | | Mycobacterium tuberculosis alanine racemase inhibited by DCS | | Descriptor: | (~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylidene-[(4~{R})-3-oxidanylidene-1,2-oxazolidin-4-yl]azanium, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | de Chiara, C, Purkiss, A, Prosser, G, Homsak, M, de Carvalho, L.P.S. | | Deposit date: | 2019-07-26 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | D-Cycloserine destruction by alanine racemase and the limit of irreversible inhibition.
Nat.Chem.Biol., 16, 2020
|
|
1H5O
 
 | | Solution structure of Crotamine, a neurotoxin from Crotalus durissus terrificus | | Descriptor: | MYOTOXIN | | Authors: | Nicastro, G, Franzoni, L, De Chiara, C, Mancin, C.A, Giglio, J.R, Spisni, A. | | Deposit date: | 2001-05-23 | | Release date: | 2003-05-09 | | Last modified: | 2013-07-24 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Crotamine, a Na+ Channel Affecting Toxin from Crotalus Durissus Terrificus Venom
Eur.J.Biochem., 270, 2003
|
|
1H8V
 
 | | The X-ray Crystal Structure of the Trichoderma reesei Family 12 Endoglucanase 3, Cel12A, at 1.9 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-BETA-1,4-GLUCANASE | | Authors: | Sandgren, M, Shaw, A, Ropp, T.H, Wu, S, Bott, R, Cameron, A.D, Stahlberg, J, Mitchinson, C, Jones, T.A. | | Deposit date: | 2001-02-16 | | Release date: | 2001-04-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-Ray Crystal Structure of the Trichoderma Reesei Family 12 Endoglucanase 3, Cel12A, at 1.9 A Resolution
J.Mol.Biol., 308, 2001
|
|
1OLQ
 
 | | The Trichoderma reesei cel12a P201C mutant, structure at 1.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-BETA-1,4-GLUCANASE | | Authors: | Sandgren, M, Gualfetti, P.J, Shaw, A, Gross, L.S, Saldajeno, M, Berglund, G.I, Jones, T.A, Mitchinson, C. | | Deposit date: | 2003-08-11 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Humicola Grisea Cel12A Enzyme Structure at 1.2 A Resolution and the Impact of its Free Cysteine Residues on Thermal Stability
Protein Sci., 12, 2003
|
|
