4JBB
 
 | | Crystal structure of Glutathione S-transferase A6TBY7(Target EFI-507184) from Klebsiella pneumoniae MGH 78578, GSH complex | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLUTATHIONE, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase A6Tby7 (Target Efi-507184) from Klebsiella Pneumoniae
To be Published
|
|
4MF6
 
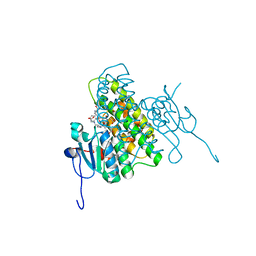 | | Crystal structure of glutathione transferase BgramDRAFT_1843 from Burkholderia graminis, Target EFI-507289, with two glutathione molecules bound per one protein subunit | | Descriptor: | BENZOIC ACID, GLUTATHIONE, Glutathione S-transferase domain | | Authors: | Patskovsky, Y, Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of glutathione transferase BgramDRAFT_1843 from Burkholderia graminis, Target EFI-507289, with two glutathione molecules bound per one protein subunit
To be Published
|
|
4JXN
 
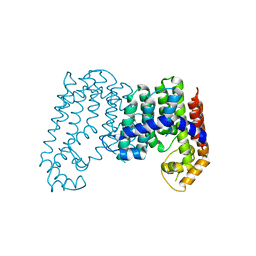 | | CRYSTAL STRUCTURE OF POLYPRENYL SYNTHASE ISP_B (TARGET EFI-509198) FROM Roseobacter denitrificans | | Descriptor: | CHLORIDE ION, Octaprenyl-diphosphate synthase, UNKNOWN LIGAND | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-03-28 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Isoprenoid Synthase Isp_B from Roseobacter Denitrificans
To be Published
|
|
4JED
 
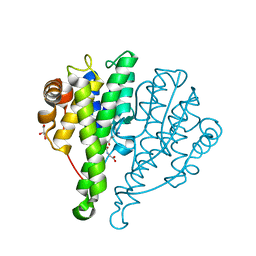 | | Crystal structure of glutathione s-transferase mrad2831_1084 (target efi-507060) from methylobacterium radiotolerans jcm 2831, complex with glutathione sulfonate | | Descriptor: | GLUTATHIONE S-TRANSFERASE, GLUTATHIONE SULFONIC ACID, GLYCEROL | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-26 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of glutathione S-transferase (TARGET EFI-507060) from Methylobacterium radiotolerans
To be Published
|
|
4MF5
 
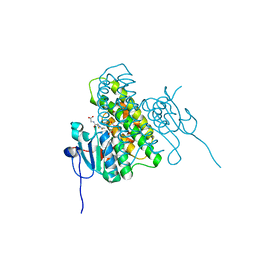 | | Crystal structure of glutathione transferase BgramDRAFT_1843 from Burkholderia graminis, Target EFI-507289, with traces of one GSH bound | | Descriptor: | FORMIC ACID, GLUTATHIONE, Glutathione S-transferase domain | | Authors: | Patskovsky, Y, Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Crystal structure of glutathione transferase BgramDRAFT_1843 from Burkholderia graminis, Target EFI-507289, with traces of one GSH bound
To be Published
|
|
4MF7
 
 | | Crystal structure of glutathione transferase BBTA-3750 from Bradyrhizobium sp., Target EFI-507290 | | Descriptor: | glutathione S-transferase enzyme with thioredoxin-like domain | | Authors: | Patskovsky, Y, Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of glutathione transferase BBTA-3750 from Bradyrhizobium sp., Target EFI-507290
To be Published
|
|
4IQG
 
 | | Crystal structure of BPRO0239 oxidoreductase from Polaromonas sp. JS666 in NADP bound form | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, GLYCEROL, ... | | Authors: | Niedzialkowska, E, Majorek, K.A, Porebski, P.J, Al Obaidi, N, Hammonds, J, Hillerich, B, Seidel, R, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-01-11 | | Release date: | 2013-01-30 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of oxidoreductase from Polaromonas sp. in NADP bound form
To be Published
|
|
4N8Y
 
 | | Crystal structure of a trap periplasmic solute binding protein from bradyrhizobium sp. btai1 b (bbta_0128), target EFI-510056 (bbta_0128), complex with alpha/beta-d-galacturonate | | Descriptor: | Putative TRAP-type C4-dicarboxylate transport system, binding periplasmic protein (DctP subunit), alpha-D-galactopyranuronic acid, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-18 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4N17
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Burkholderia ambifaria (BAM_6123), Target EFI-510059, With bound beta-D-galacturonate | | Descriptor: | CALCIUM ION, CHLORIDE ION, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Zhao, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-03 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4N6K
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Desulfovibrio salexigens DSM2638, Target EFI-510113 (Desal_0342), complex with diglycerolphosphate | | Descriptor: | TRAP dicarboxylate transporter-DctP subunit, bis[(2S)-2,3-dihydroxypropyl] hydrogen phosphate | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-13 | | Release date: | 2013-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4N91
 
 | | Crystal structure of a trap periplasmic solute binding protein from anaerococcus prevotii dsm 20548 (Apre_1383), target EFI-510023, with bound alpha/beta d-glucuronate | | Descriptor: | CHLORIDE ION, TETRAETHYLENE GLYCOL, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-18 | | Release date: | 2013-11-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4MX6
 
 | | Crystal structure of a trap periplasmic solute binding protein from shewanella oneidensis (SO_3134), target EFI-510275, with bound succinate | | Descriptor: | CHLORIDE ION, SUCCINIC ACID, TRAP-type C4-dicarboxylate:H+ symport system substrate-binding component DctP | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-25 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4N6D
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Desulfovibrio salexigens DSM2638 (Desal_3247), Target EFI-510112, phased with I3C, open complex, C-terminus of symmetry mate bound in ligand binding site | | Descriptor: | 1,2-ETHANEDIOL, 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-11 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4NAP
 
 | | Crystal structure of a trap periplasmic solute binding protein from Desulfovibrio alaskensis G20 (DDE_0634), target EFI-510102, with bound d-tryptophan | | Descriptor: | D-TRYPTOPHAN, Extracellular solute-binding protein, family 7 | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-13 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4N8G
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Chromohalobacter salexigens DSM 3043 (Csal_0660), Target EFI-501075, with bound D-alanine-D-alanine | | Descriptor: | CHLORIDE ION, D-ALANINE, SULFATE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4NGU
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Desulfovibrio alaskensis G20 (Dde_1548), Target EFI-510103, with bound D-Ala-D-Ala | | Descriptor: | CHLORIDE ION, D-ALANINE, SULFATE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-11-02 | | Release date: | 2013-12-04 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4NHB
 
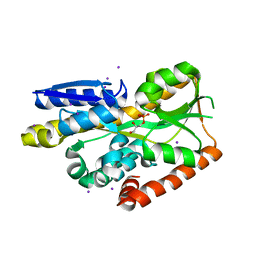 | | Crystal structure of a TRAP periplasmic solute binding protein from Desulfovibrio desulfuricans (Ddes_1525), Target EFI-510107, with bound sn-glycerol-3-phosphate | | Descriptor: | IODIDE ION, SN-GLYCEROL-3-PHOSPHATE, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-11-04 | | Release date: | 2013-11-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4NF0
 
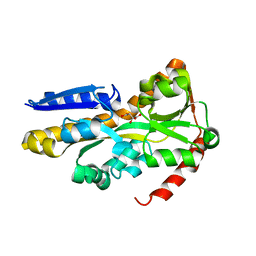 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM PSEUDOMONAS AERUGINOSA PAO1 (PA4616), TARGET EFI-510182, WITH BOUND L-Malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Probable c4-dicarboxylate-binding protein, SULFATE ION | | Authors: | Vetting, M.W, Patskovsky, Y, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-30 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4NG7
 
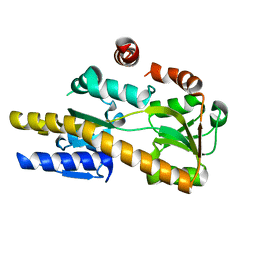 | | Crystal structure of a TRAP periplasmic solute binding protein from Citrobacter koseri (CKO_04899), Target EFI-510094, apo, open structure | | Descriptor: | TRAP periplasmic solute binding protein | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-11-01 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
3V4B
 
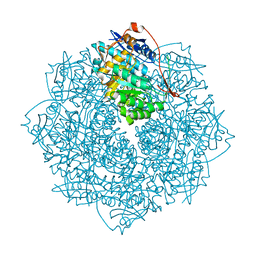 | | Crystal structure of an enolase from the soil bacterium Cellvibrio japonicus (TARGET EFI-502161) with bound MG and L-tartrate | | Descriptor: | CHLORIDE ION, L(+)-TARTARIC ACID, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Seidel, R.D, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Al Obaidi, N, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-12-14 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of an enolase from the soil bacterium Cellvibrio japonicus (TARGET EFI-502161) with bound MG and l-tartrate
to be published
|
|
4UAB
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Chromohalobacter salexigens DSM 3043 (Csal_0678), Target EFI-501078, with bound ethanolamine | | Descriptor: | CHLORIDE ION, ETHANOLAMINE, Twin-arginine translocation pathway signal | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-08-08 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4Y9T
 
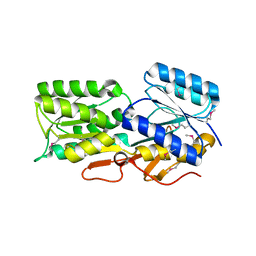 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS S4 (Avi_5305, TARGET EFI-511224) WITH BOUND ALPHA-D-GLUCOSAMINE | | Descriptor: | 2-amino-2-deoxy-alpha-D-glucopyranose, ABC transporter, solute binding protein | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-02-17 | | Release date: | 2015-03-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure of an ABC transporter solute-binding protein specific for the amino sugars glucosamine and galactosamine.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5BR1
 
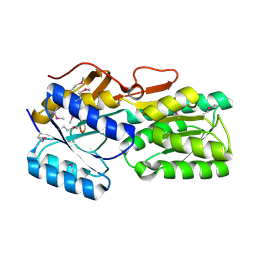 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS S4 (Avi_5305, TARGET EFI-511224) WITH BOUND ALPHA-D-GALACTOSAMINE | | Descriptor: | 2-amino-2-deoxy-alpha-D-galactopyranose, ABC transporter, binding protein | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-05-29 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of an ABC transporter solute-binding protein specific for the amino sugars glucosamine and galactosamine.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6CHK
 
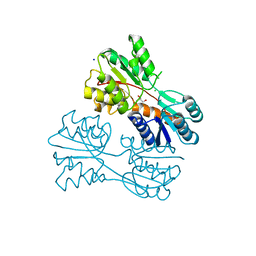 | | Crystal structure of LacI family transcriptional regulator from Lactobacillus casei, Target EFI-512911, with bound TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, SODIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Shabalin, I.G, Kowiel, M, Porebski, P.J, Minor, W, Jaskolski, M, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative, E.F.I. | | Deposit date: | 2018-02-22 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Automatic recognition of ligands in electron density by machine learning.
Bioinformatics, 35, 2019
|
|
4NX1
 
 | | Crystal structure of a trap periplasmic solute binding protein from Sulfitobacter sp. nas-14.1, target EFI-510292, with bound alpha-D-taluronate | | Descriptor: | C4-dicarboxylate transport system substrate-binding protein, alpha-D-talopyranuronic acid | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-08 | | Release date: | 2014-01-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
