4GEW
 
 | |
4FVA
 
 | | Crystal structure of truncated Caenorhabditis elegans TDP2 | | Descriptor: | 1,2-ETHANEDIOL, 5'-tyrosyl-DNA phosphodiesterase, MAGNESIUM ION, ... | | Authors: | Shi, K, Kurahashi, K, Aihara, H. | | Deposit date: | 2012-06-29 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis for recognition of 5'-phosphotyrosine adducts by Tdp2.
Nat.Struct.Mol.Biol., 19, 2012
|
|
5SXG
 
 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B | | Descriptor: | 1,3-PROPANDIOL, DNA dC->dU-editing enzyme APOBEC-3B, IMIDAZOLE, ... | | Authors: | Shi, K, Kurahashi, K, Aihara, H. | | Deposit date: | 2016-08-09 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Conformational Switch Regulates the DNA Cytosine Deaminase Activity of Human APOBEC3B.
Sci Rep, 7, 2017
|
|
5SWW
 
 | | Crystal Structure of Human APOBEC3A complexed with ssDNA | | Descriptor: | DNA 15-Mer, DNA dC->dU-editing enzyme APOBEC-3A, GLYCEROL, ... | | Authors: | Shi, K, Banerjee, S, Kurahashi, K, Aihara, H. | | Deposit date: | 2016-08-09 | | Release date: | 2016-12-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.151 Å) | | Cite: | Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7U0N
 
 | | Crystal structure of chimeric omicron RBD (strain BA.1) complexed with human ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Geng, Q, Shi, K, Ye, G, Zhang, W, Aihara, H, Li, F. | | Deposit date: | 2022-02-18 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural Basis for Human Receptor Recognition by SARS-CoV-2 Omicron Variant BA.1.
J.Virol., 96, 2022
|
|
6XC1
 
 | | Crystal structure of bacteriophage T4 spackle and lysozyme in orthorhombic form | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, Lysozyme, ... | | Authors: | Shi, K, Oakland, J.T, Kurniawan, F, Moeller, N.H, Aihara, H. | | Deposit date: | 2020-06-07 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis of superinfection exclusion by bacteriophage T4 Spackle.
Commun Biol, 3, 2020
|
|
7MC5
 
 | | Crystal structure of the SARS-CoV-2 ExoN-nsp10 complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, L(+)-TARTARIC ACID, ... | | Authors: | Moeller, N.M, Shi, K, Banerjee, S, Yin, L, Aihara, H. | | Deposit date: | 2021-04-01 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6XC0
 
 | | Crystal structure of bacteriophage T4 spackle and lysozyme in monoclinic form | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Shi, K, Oakland, J.T, Kurniawan, F, Moeller, N.H, Aihara, H. | | Deposit date: | 2020-06-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural basis of superinfection exclusion by bacteriophage T4 Spackle.
Commun Biol, 3, 2020
|
|
5CQK
 
 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3B, GLYCEROL, SODIUM ION, ... | | Authors: | Shi, K, Kurahashi, K, Aihara, H. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of the DNA Deaminase APOBEC3B Catalytic Domain.
J.Biol.Chem., 290, 2015
|
|
5CQI
 
 | |
5CQH
 
 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Shi, K, Kurahashi, K, Aihara, H. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of the DNA Deaminase APOBEC3B Catalytic Domain.
J.Biol.Chem., 290, 2015
|
|
5U0M
 
 | | Fatty aldehyde dehydrogenase from Marinobacter aquaeolei VT8 and cofactor complex | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, N-succinylglutamate 5-semialdehyde dehydrogenase, ... | | Authors: | Shi, K, Mulliner, K, Barney, B.M, Aihara, H. | | Deposit date: | 2016-11-24 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.075 Å) | | Cite: | Five Fatty Aldehyde Dehydrogenase Enzymes from Marinobacter and Acinetobacter spp. and Structural Insights into the Aldehyde Binding Pocket.
Appl. Environ. Microbiol., 83, 2017
|
|
7R9C
 
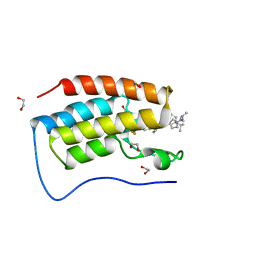 | | Cocrystal of BRD4(D1) with N,N-dimethyl-2-[(3R)-3-(5-{2-[2-methyl-5-(propan-2-yl)phenoxy]pyrimidin-4-yl}-4-[4-(trifluoromethyl)phenyl]-1H-imidazol-1-yl)pyrrolidin-1-yl]ethan-1-amine | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, CHLORIDE ION, ... | | Authors: | Cui, H, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
7UFK
 
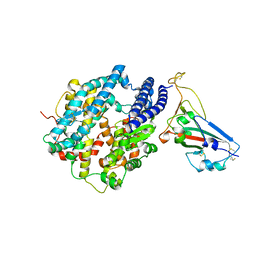 | | Crystal structure of chimeric omicron RBD (strain BA.2) complexed with human ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, W, Shi, K, Geng, Q, Ye, G, Aihara, H, Li, F. | | Deposit date: | 2022-03-22 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural basis for mouse receptor recognition by SARS-CoV-2 omicron variant.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UFL
 
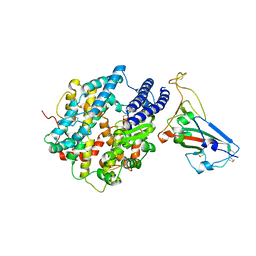 | | Crystal structure of chimeric omicron RBD (strain BA.2) complexed with chimeric mouse ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, W, Shi, K, Geng, Q, Ye, G, Aihara, H, Li, F. | | Deposit date: | 2022-03-22 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structural basis for mouse receptor recognition by SARS-CoV-2 omicron variant.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5CQD
 
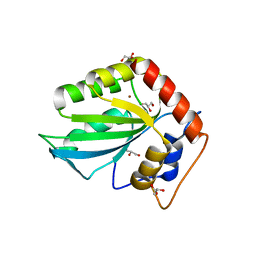 | |
4FPV
 
 | | Crystal structure of D. rerio TDP2 complexed with single strand DNA product | | Descriptor: | DNA (5'-D(P*TP*GP*CP*AP*G)-3'), GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shi, K, Kurahashi, K, Aihara, H. | | Deposit date: | 2012-06-22 | | Release date: | 2012-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural basis for recognition of 5'-phosphotyrosine adducts by Tdp2.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4FW2
 
 | |
5TD5
 
 | | Crystal Structure of Human APOBEC3B variant complexed with ssDNA | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(P*TP*TP*CP*AP*T)-3'), ... | | Authors: | Shi, K, Banerjee, S, Kurahashi, K, Aihara, H. | | Deposit date: | 2016-09-16 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.718 Å) | | Cite: | Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6X6O
 
 | | Crystal structure of T4 protein Spackle as determined by native SAD phasing | | Descriptor: | CHLORIDE ION, Protein spackle | | Authors: | Shi, K, Kurniawan, F, Banerjee, S, Moeller, N.H, Aihara, H. | | Deposit date: | 2020-05-28 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structure of bacteriophage T4 Spackle as determined by native SAD phasing.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5SXH
 
 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B | | Descriptor: | 1,2-ETHANEDIOL, DNA dC->dU-editing enzyme APOBEC-3B, ZINC ION | | Authors: | Shi, K, Kurahashi, K, Aihara, H. | | Deposit date: | 2016-08-09 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Conformational Switch Regulates the DNA Cytosine Deaminase Activity of Human APOBEC3B.
Sci Rep, 7, 2017
|
|
7RXS
 
 | | Crystal of BRD4(D1) with 2-[(3S)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3S)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine, Bromodomain-containing protein 4 | | Authors: | Cui, H, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
7RXT
 
 | | Crystal of BRD4(D1) with 2-[(3R)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine | | Descriptor: | 2-[(3R)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine, Bromodomain-containing protein 4 | | Authors: | Cui, H, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
7RXR
 
 | | Crystal Structure of BRD4(D1) with 4-[4-(4-bromophenyl)-1-(piperidin-4-yl)-1H-imidazol-5-yl]-N-(3,5-dimethylphenyl)pyrimidin-2-amine | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-(4-bromophenyl)-1-(piperidin-4-yl)-1H-imidazol-5-yl]-N-(3,5-dimethylphenyl)pyrimidin-2-amine, Bromodomain-containing protein 4 | | Authors: | Cui, H, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
5U0L
 
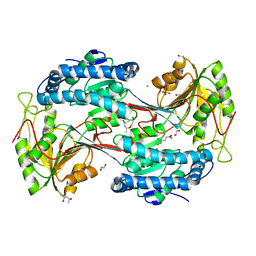 | | X-ray crystal structure of fatty aldehyde dehydrogenase enzymes from Marinobacter aquaeolei VT8 complexed with a substrate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Shi, K, Mulliner, K, Barney, B.M, Aihara, H. | | Deposit date: | 2016-11-24 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Five Fatty Aldehyde Dehydrogenase Enzymes from Marinobacter and Acinetobacter spp. and Structural Insights into the Aldehyde Binding Pocket.
Appl. Environ. Microbiol., 83, 2017
|
|
