3HVA
 
 | |
8Z3F
 
 | | Complex structure of CIB2 and TMC1 CR1 | | Descriptor: | CALCIUM ION, Calcium and integrin-binding family member 2, Transmembrane channel-like protein 1 | | Authors: | Wu, S, Lin, L, Lu, Q. | | Deposit date: | 2024-04-15 | | Release date: | 2025-02-19 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Mechano-electrical transduction components TMC1-CIB2 undergo a Ca 2+ -induced conformational change linked to hearing loss.
Dev.Cell, 60, 2025
|
|
8WHA
 
 | | Structure of DDM1-nucleosome complex in the ADP-BeFx state with DDM1 bound to SHL2 and SHL-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WH5
 
 | | Structure of DDM1-nucleosome complex in the apo state | | Descriptor: | ATP-dependent DNA helicase DDM1, DNA (antisense strand), DNA (sense strand), ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WHB
 
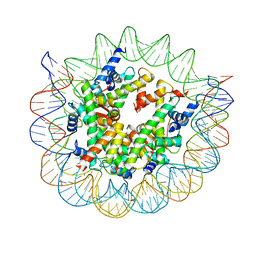 | | Structure of nucleosome core particle of Arabidopsis thaliana | | Descriptor: | DNA (antisense strand), DNA (sense strand), Histone H2A.6, ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-23 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WH8
 
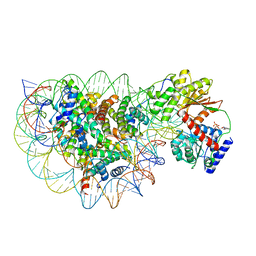 | | Structure of DDM1-nucleosome complex in ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, DNA (antisense strand), ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WH9
 
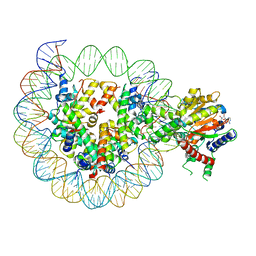 | | Structure of DDM1-nucleosome complex in ADP-BeFx state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
7DWO
 
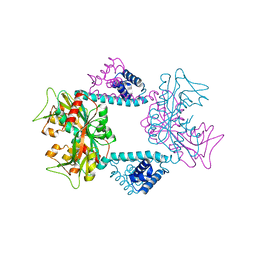 | |
7DWN
 
 | | Crystal structure of Vibrio fischeri DarR in complex with DNA reveals the transcriptional activation mechanism of LTTR family members | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Predicted DNA-binding transcriptional regulator | | Authors: | Wang, W.W, Wu, H, He, J.H, Yu, F. | | Deposit date: | 2021-01-17 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structure details of Vibrio fischeri DarR and mutant DarR-M202I from LTTR family reveals their activation mechanism.
Int.J.Biol.Macromol., 183, 2021
|
|
8WEC
 
 | | Crystal structure of Arabidopsis thaliana MIK2 ectodomain in complex with BAK1 ectodomain and SCOOP12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wan, L.H, Hu, Y.X, Wu, H.M. | | Deposit date: | 2023-09-17 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanistic study of SCOOPs recognition by MIK2-BAK1 complex reveals the role of N-glycans in plant ligand-receptor-coreceptor complex formation.
Nat.Plants, 10, 2024
|
|
8WEB
 
 | | Crystal structure of Arabidopsis thaliana MIK2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MDIS1-interacting receptor like kinase 2 | | Authors: | Wan, L.H, Hu, Y.X, Wu, H.M. | | Deposit date: | 2023-09-17 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanistic study of SCOOPs recognition by MIK2-BAK1 complex reveals the role of N-glycans in plant ligand-receptor-coreceptor complex formation.
Nat.Plants, 10, 2024
|
|
8WED
 
 | | Crystal structure of Arabidopsis thaliana MIK2 ectodomain in complex with BAK1 ectodomain and Fusarium oxysporum SCOOPL | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wan, L.H, Hu, Y.X, Wu, H.M. | | Deposit date: | 2023-09-17 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Mechanistic study of SCOOPs recognition by MIK2-BAK1 complex reveals the role of N-glycans in plant ligand-receptor-coreceptor complex formation.
Nat.Plants, 10, 2024
|
|
8WEF
 
 | | Crystal structure of Brassica napus MIK2 ectodomain in complex with Fusarium oxysporum SCOOPL | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wan, L.H, Hu, Y.X, Wu, H.M. | | Deposit date: | 2023-09-17 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic study of SCOOPs recognition by MIK2-BAK1 complex reveals the role of N-glycans in plant ligand-receptor-coreceptor complex formation.
Nat.Plants, 10, 2024
|
|
8WEI
 
 | | Crystal structure of Brassica napus MIK2 ectodomain (N393D mutant) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wan, L.H, Hu, Y.X, Wu, H.M. | | Deposit date: | 2023-09-17 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Mechanistic study of SCOOPs recognition by MIK2-BAK1 complex reveals the role of N-glycans in plant ligand-receptor-coreceptor complex formation.
Nat.Plants, 10, 2024
|
|
8WEG
 
 | | Crystal structure of Brassica napus MIK2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wan, L.H, Hu, Y.X, Wu, H.M. | | Deposit date: | 2023-09-17 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Mechanistic study of SCOOPs recognition by MIK2-BAK1 complex reveals the role of N-glycans in plant ligand-receptor-coreceptor complex formation.
Nat.Plants, 10, 2024
|
|
3HV8
 
 | | Crystal structure of FimX EAL domain from Pseudomonas aeruginosa bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Protein FimX | | Authors: | Navarro, M.V.A.S, De, N, Bae, N, Sondermann, H. | | Deposit date: | 2009-06-15 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.445 Å) | | Cite: | Structural analysis of the GGDEF-EAL domain-containing c-di-GMP receptor FimX.
Structure, 17, 2009
|
|
8WEH
 
 | | Crystal structure of Brassica napus MIK2 ectodomain (N393A mutant) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wan, L.H, Hu, Y.X, Wu, H.M. | | Deposit date: | 2023-09-17 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Mechanistic study of SCOOPs recognition by MIK2-BAK1 complex reveals the role of N-glycans in plant ligand-receptor-coreceptor complex formation.
Nat.Plants, 10, 2024
|
|
8WEE
 
 | | Crystal structure of Arabidopsis thaliana MIK2 ectodomain in complex with SCOOP12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wan, L.H, Hu, Y.X, Wu, H.M. | | Deposit date: | 2023-09-17 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Mechanistic study of SCOOPs recognition by MIK2-BAK1 complex reveals the role of N-glycans in plant ligand-receptor-coreceptor complex formation.
Nat.Plants, 10, 2024
|
|
7EO8
 
 | | Crystal structure of SARS coronavirus main protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2808516 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7CJF
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody heavy chain, ... | | Authors: | Guo, Y, Li, X, Zhang, G, Fu, D, Schweizer, L, Zhang, H, Rao, Z. | | Deposit date: | 2020-07-10 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | A SARS-CoV-2 neutralizing antibody with extensive Spike binding coverage and modified for optimal therapeutic outcomes.
Nat Commun, 12, 2021
|
|
7DQZ
 
 | | Crystal structure of SARS 3C-like protease in apo form | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-12-24 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7C8D
 
 | | Cryo-EM structure of cat ACE2 and SARS-CoV-2 RBD | | Descriptor: | Angiotensin-converting enzyme 2, Spike protein S1, ZINC ION | | Authors: | Gao, G.F, Wang, Q.H, Wu, L.l. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-02 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Broad host range of SARS-CoV-2 and the molecular basis for SARS-CoV-2 binding to cat ACE2.
Cell Discov, 6, 2020
|
|
7QUO
 
 | | FimH lectin domain in complex with oligomannose-6 | | Descriptor: | FimH, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Bouckaert, J, Bourenkov, G.P. | | Deposit date: | 2022-01-18 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into a cooperative switch between one and two FimH bacterial adhesins binding pauci- and high-mannose type N-glycan receptors
J.Biol.Chem., 299, 2023
|
|
9BLL
 
 | | Cryo-EM of RBD(EG5.1)/1301B7 Fab Complex | | Descriptor: | 1301B7 Heavy Chain, 1301B7 Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, M.R, Green, T.J. | | Deposit date: | 2024-04-30 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Potent neutralization by a RBD antibody with broad specificity for SARS-CoV-2 JN.1 and other variants.
Npj Viruses, 2, 2024
|
|
7WB4
 
 | | Cryo-EM structure of the NR subunit from X. laevis NPC | | Descriptor: | GATOR complex protein SEC13, MGC154553 protein, MGC83295 protein, ... | | Authors: | Huang, G, Zhan, X, Shi, Y. | | Deposit date: | 2021-12-15 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Cryo-EM structure of the nuclear ring from Xenopus laevis nuclear pore complex.
Cell Res., 32, 2022
|
|
