7WNB
 
 | |
3A71
 
 | |
5FVF
 
 | | Room temperature structure of IrisFP determined by serial femtosecond crystallography. | | Descriptor: | AMMONIUM ION, Green to red photoconvertible GFP-like protein EosFP, SULFATE ION | | Authors: | Colletier, J.P, Gallat, F.X, Coquelle, N, Weik, M. | | Deposit date: | 2016-02-06 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Serial Femtosecond Crystallography and Ultrafast Absorption Spectroscopy of the Photoswitchable Fluorescent Protein Irisfp.
J.Phys.Chem.Lett., 7, 2016
|
|
5FVI
 
 | | Structure of IrisFP in mineral grease at 100 K. | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION | | Authors: | Colletier, J.P, Gallat, F.X, Coquelle, N, Weik, M. | | Deposit date: | 2016-02-07 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Serial Femtosecond Crystallography and Ultrafast Absorption Spectroscopy of the Photoswitchable Fluorescent Protein Irisfp.
J.Phys.Chem.Lett., 7, 2016
|
|
3A72
 
 | |
7DKJ
 
 | |
6IWH
 
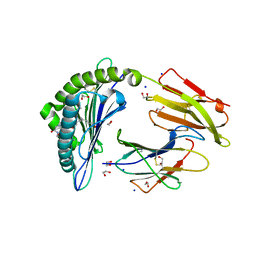 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*05104 complexed with C14-GGGI lipopeptide | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, C14-GGGI lipopeptide, ... | | Authors: | Yamamoto, Y, Morita, D, Sugita, M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification and Structure of an MHC Class I-Encoded Protein with the Potential to PresentN-Myristoylated 4-mer Peptides to T Cells.
J Immunol., 202, 2019
|
|
6IWG
 
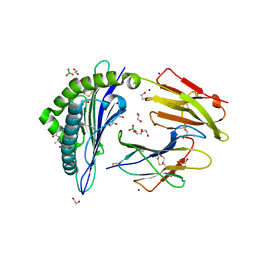 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*05104 complexed with N-myristoylated 4-mer lipopeptide derived from SIV nef protein | | Descriptor: | 1,2-ETHANEDIOL, BORIC ACID, Beta-2-microglobulin, ... | | Authors: | Yamamoto, Y, Morita, D, Sugita, M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and Structure of an MHC Class I-Encoded Protein with the Potential to PresentN-Myristoylated 4-mer Peptides to T Cells.
J Immunol., 202, 2019
|
|
1FA3
 
 | |
3CLN
 
 | |
4M7I
 
 | | Crystal Structure of GSK6157 Bound to PERK (R587-R1092, delete A660-T867) at 2.34A Resolution | | Descriptor: | 1-[5-(4-amino-7-methyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)-4-fluoro-1H-indol-1-yl]-2-(6-methylpyridin-2-yl)ethanone, Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Gampe, R.T, Axten, J.M. | | Deposit date: | 2013-08-12 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Discovery of 5-{4-fluoro-1-[(6-methyl-2-pyridinyl)acetyl]-2,3-dihydro-1H-indol-5-yl}-7-methyl-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2656157), a Potent and Selective PERK Inhibitor Selected for Preclinical Development
To be Published
|
|
7FGL
 
 | | The complex structure of PHF core domain peptide of tau, VQIVYK, and antibody's Fab domain. | | Descriptor: | AMMONIUM ION, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Tsuchida, T, Tsuchiya, T, Miyamoto, K, In, Y, Minoura, K, Taniguchi, T, Ishida, T, Tomoo, K. | | Deposit date: | 2021-07-27 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The cross-reaction complex structure with VQIVYK of tau and the antibody's Fab domain.
To Be Published
|
|
5X7Y
 
 | | Crystal Structure of the Dog Lipocalin Allergen Can f 6 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Lipocalin-Can f 6 allergen | | Authors: | Yamamoto, K, Otani, T, Sugiura, K, Nakatsuji, M, Nishimura, S, Inui, T. | | Deposit date: | 2017-02-28 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the dog allergen Can f 6 and structure-based implications of its cross-reactivity with the cat allergen Fel d 4.
Sci Rep, 9, 2019
|
|
6JIG
 
 | | Crystal structure of GMP reductase C318A from Trypanosoma brucei in complex with guanosine 5'-monophosphate | | Descriptor: | GMP reductase, GUANOSINE-5'-MONOPHOSPHATE, POTASSIUM ION | | Authors: | Mase, H, Imamura, A, Nishimura, S, Inui, T. | | Deposit date: | 2019-02-21 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Allosteric regulation accompanied by oligomeric state changes of Trypanosoma brucei GMP reductase through cystathionine-beta-synthase domain.
Nat Commun, 11, 2020
|
|
6JL8
 
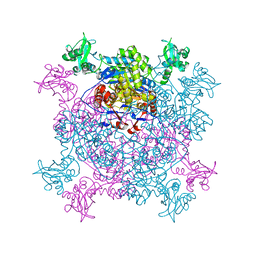 | |
3AU5
 
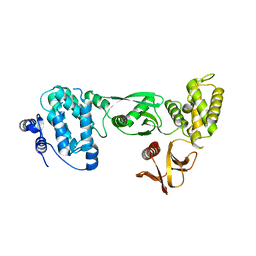 | |
7VPY
 
 | | Crystal structure of the neutralizing nanobody P86 against SARS-CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Nanobody, SULFATE ION | | Authors: | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
7VQ0
 
 | | Cryo-EM structure of the SARS-CoV-2 spike protein (2-up RBD) bound to neutralizing nanobodies P86 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
6KAK
 
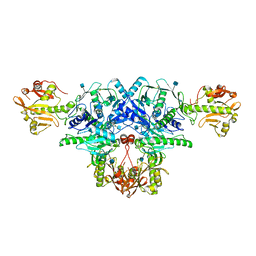 | | Crystal structure of FKRP in complex with Mg ion | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fukutin-related protein, MAGNESIUM ION, ... | | Authors: | Kuwabara, N. | | Deposit date: | 2019-06-23 | | Release date: | 2020-01-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.056 Å) | | Cite: | Crystal structures of fukutin-related protein (FKRP), a ribitol-phosphate transferase related to muscular dystrophy.
Nat Commun, 11, 2020
|
|
6KAM
 
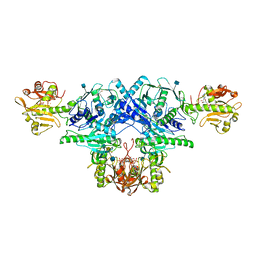 | | Crystal structure of FKRP in complex with Ba ion, CDP-ribtol, and sugar acceptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, BARIUM ION, ... | | Authors: | Kuwabara, N. | | Deposit date: | 2019-06-23 | | Release date: | 2020-01-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal structures of fukutin-related protein (FKRP), a ribitol-phosphate transferase related to muscular dystrophy.
Nat Commun, 11, 2020
|
|
6KAJ
 
 | | Crystal structure of FKRP in complex with Ba ion | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BARIUM ION, CYTIDINE-5'-DIPHOSPHATE, ... | | Authors: | Kuwabara, N. | | Deposit date: | 2019-06-23 | | Release date: | 2020-01-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2249 Å) | | Cite: | Crystal structures of fukutin-related protein (FKRP), a ribitol-phosphate transferase related to muscular dystrophy.
Nat Commun, 11, 2020
|
|
2Q2Z
 
 | | Crystal Structure of KSP in Complex with Inhibitor 22 | | Descriptor: | 1-[(4R)-4-[3-(4-ACETYLPIPERAZIN-1-YL)PROPYL]-1-(2-FLUORO-5-METHYLPHENYL)-4-PHENYL-4,5-DIHYDRO-1H-PYRAZOL-3-YL]ETHANONE, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yan, Y. | | Deposit date: | 2007-05-29 | | Release date: | 2007-09-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Kinesin spindle protein (KSP) inhibitors. Part 8: Design and synthesis of 1,4-diaryl-4,5-dihydropyrazoles as potent inhibitors of the mitotic kinesin KSP.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
6KAL
 
 | | Crystal structure of FKRP in complex with Mg ion and CMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYTIDINE-5'-MONOPHOSPHATE, Fukutin-related protein, ... | | Authors: | Kuwabara, N. | | Deposit date: | 2019-06-23 | | Release date: | 2020-01-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of fukutin-related protein (FKRP), a ribitol-phosphate transferase related to muscular dystrophy.
Nat Commun, 11, 2020
|
|
6KAN
 
 | | Crystal structure of FKRP in complex with Ba ion | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BARIUM ION, Fukutin-related protein, ... | | Authors: | Kuwabara, N. | | Deposit date: | 2019-06-23 | | Release date: | 2020-01-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Crystal structures of fukutin-related protein (FKRP), a ribitol-phosphate transferase related to muscular dystrophy.
Nat Commun, 11, 2020
|
|
3A7E
 
 | | Crystal structure of human COMT complexed with SAM and 3,5-dinitrocatechol | | Descriptor: | 3,5-DINITROCATECHOL, Catechol O-methyltransferase, MAGNESIUM ION, ... | | Authors: | Tsuji, E. | | Deposit date: | 2009-09-26 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Hit to Lead: Comprehensive Strategy of de novo Scaffold Generation by FBDD. Part 1: In silico Fragments Linking and Verification of Spatial Proximity using Inter Ligand NOE Approachs
To be Published
|
|
