8J8V
 
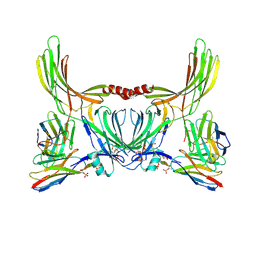 | | Structure of beta-arrestin2 in complex with D6Rpp (Local Refine) | | Descriptor: | Atypical chemokine receptor 2, Beta-arrestin-2, Fab30 Heavy Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-05-02 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Molecular insights into atypical modes of beta-arrestin interaction with seven transmembrane receptors.
Science, 383, 2024
|
|
7FF6
 
 | | The 0.83 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with cis-vaccenic acid | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-22 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.83 Å) | | Cite: | The 0.83 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with cis-vaccenic acid
To Be Published
|
|
7FEK
 
 | | The 1.05 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with perfluorooctanoic acid | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Hara, T, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-21 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The 1.05 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with perfluorooctanoic acid
To Be Published
|
|
8JA3
 
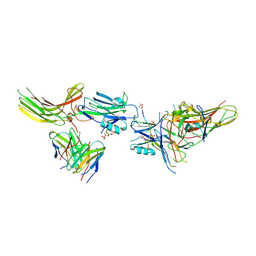 | | Structure of beta-arrestin1 in complex with C3aRpp | | Descriptor: | Beta-arrestin-1, C3a anaphylatoxin chemotactic receptor, Fab30 heavy chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-05-05 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Molecular insights into atypical modes of beta-arrestin interaction with seven transmembrane receptors.
Science, 383, 2024
|
|
8J9K
 
 | | Structure of basal beta-arrestin2 | | Descriptor: | Beta-arrestin-2, Fab6 heavy chain, Fab6 light chain | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-05-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular insights into atypical modes of beta-arrestin interaction with seven transmembrane receptors.
Science, 383, 2024
|
|
1TN3
 
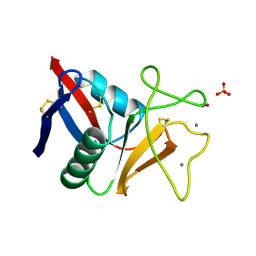 | | THE C-TYPE LECTIN CARBOHYDRATE RECOGNITION DOMAIN OF HUMAN TETRANECTIN | | Descriptor: | CALCIUM ION, ETHANOL, SULFATE ION, ... | | Authors: | Kastrup, J.S, Nielsen, B.B, Rasmussen, H, Holtet, T.L, Graversen, J.H, Etzerodt, M, Thoegersen, H.C, Larsen, I.K. | | Deposit date: | 1997-11-06 | | Release date: | 1998-05-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the C-type lectin carbohydrate recognition domain of human tetranectin.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
7FFX
 
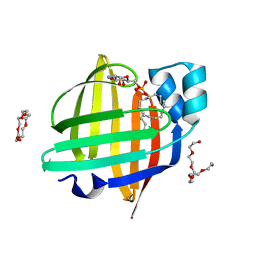 | | The 0.88 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with alpha-llinolenic acid | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, ALPHA-LINOLENIC ACID, Fatty acid-binding protein, ... | | Authors: | Sugiyama, S, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-24 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | The 0.88 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with alpha-llinolenic acid
To Be Published
|
|
7FG1
 
 | | The 0.93 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with tridecanoic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Takahashi, J, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | The 0.93 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with tridecanoic acid
To Be Published
|
|
7FG5
 
 | | Room temperature structure of the human heart fatty acid-binding protein complexed with capric acid | | Descriptor: | DECANOIC ACID, Fatty acid-binding protein, heart | | Authors: | Sugiyama, S, Kakinouchi, K, Takahashi, J, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-26 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Room temperature structure of the human heart fatty acid-binding protein complexed with capric acid
To Be Published
|
|
7U97
 
 | | SAAV pH 4.0 capsid structure | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2022-03-10 | | Release date: | 2022-04-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Characterization of the Serpentine Adeno-Associated Virus (SAAV) Capsid Structure: Receptor Interactions and Antigenicity.
J.Virol., 96, 2022
|
|
7U95
 
 | | SAAV pH 6.0 capsid structure | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2022-03-10 | | Release date: | 2022-04-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Characterization of the Serpentine Adeno-Associated Virus (SAAV) Capsid Structure: Receptor Interactions and Antigenicity.
J.Virol., 96, 2022
|
|
7U96
 
 | | SAAV pH 5.5 capsid structure | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2022-03-10 | | Release date: | 2022-04-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Characterization of the Serpentine Adeno-Associated Virus (SAAV) Capsid Structure: Receptor Interactions and Antigenicity.
J.Virol., 96, 2022
|
|
7U94
 
 | | SAAV pH 7.4 capsid structure | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2022-03-10 | | Release date: | 2022-04-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Characterization of the Serpentine Adeno-Associated Virus (SAAV) Capsid Structure: Receptor Interactions and Antigenicity.
J.Virol., 96, 2022
|
|
1TNM
 
 | |
1KBQ
 
 | | Complex of Human NAD(P)H quinone Oxidoreductase with 5-methoxy-1,2-dimethyl-3-(4-nitrophenoxymethyl)indole-4,7-dione (ES936) | | Descriptor: | 5-METHOXY-1,2-DIMETHYL-3-(4-NITROPHENOXYMETHYL)INDOLE-4,7-DIONE, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Faig, M, Bianchet, M.A, Amzel, L.M. | | Deposit date: | 2001-11-06 | | Release date: | 2002-01-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of a mechanism-based inhibitor of NAD(P)H:quinone oxidoreductase 1 by biochemical, X-ray crystallographic, and mass spectrometric approaches.
Biochemistry, 40, 2001
|
|
6ZJU
 
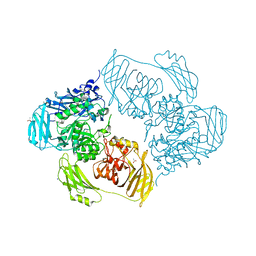 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E441Q in complex with saccharose | | Descriptor: | ACETATE ION, Beta-galactosidase, MALONATE ION, ... | | Authors: | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mapping the Transglycosylation Relevant Sites of Cold-Adapted beta-d-Galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 21, 2020
|
|
5FTW
 
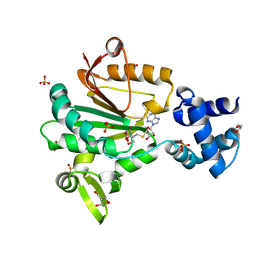 | | Crystal structure of glutamate O-methyltransferase in complex with S- adenosyl-L-homocysteine (SAH) from Bacillus subtilis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHEMOTAXIS PROTEIN METHYLTRANSFERASE, GLYCEROL, ... | | Authors: | Sharma, R, Dhindwal, S, Batra, M, Aggarwal, M, Kumar, P, Tomar, S. | | Deposit date: | 2016-01-18 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Pentapeptide-Independent Chemotaxis Receptor Methyltransferase (Cher) Reveals Idiosyncratic Structural Determinants for Receptor Recognition.
J.Struct.Biol., 196, 2016
|
|
5M7Z
 
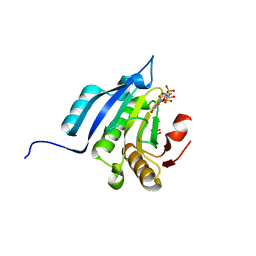 | | Translation initiation factor 4E in complex with (SP)-m2(7,2'O)GppSepG mRNA 5' cap analog (beta-Se-ARCA D2) | | Descriptor: | CHLORIDE ION, Eukaryotic translation initiation factor 4E, GLYCEROL, ... | | Authors: | Warminski, M, Nowak, E, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2016-10-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.689 Å) | | Cite: | Translation initiation factor 4E in complex with (SP)-m2(7,2'O)GppSepG mRNA 5' cap analog (beta-Se-ARCA D2)
To Be Published
|
|
5ML5
 
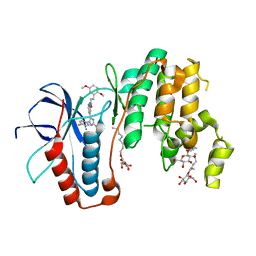 | | Human p38alpha MAPK in complex with imidazolyl pyridine inhibitor 11b | | Descriptor: | 3-(2,5-dimethoxyphenyl)-~{N}-[4-[4-(4-fluorophenyl)-2-methylsulfanyl-1~{H}-imidazol-5-yl]pyridin-2-yl]propanamide, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Buehrmann, M, Rauh, D. | | Deposit date: | 2016-12-06 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimized 4,5-Diarylimidazoles as Potent/Selective Inhibitors of Protein Kinase CK1 delta and Their Structural Relation to p38 alpha MAPK.
Molecules, 22, 2017
|
|
8IY9
 
 | | Structure of Niacin-GPR109A-G protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(o) subunit alpha, ... | | Authors: | Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-04-04 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution.
Nat Commun, 15, 2024
|
|
1KBG
 
 | | MHC Class I H-2KB Presented Glycopeptide RGY8-6H-GAL2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (BETA-2-MICROGLOBULIN), ... | | Authors: | Speir, J.A, Abdel-Motal, U.M, Jondal, M, Wilson, I.A. | | Deposit date: | 1998-08-28 | | Release date: | 1999-02-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an MHC class I presented glycopeptide that generates carbohydrate-specific CTL.
Immunity, 10, 1999
|
|
1UB3
 
 | | Crystal Structure of Tetrameric Structure of Aldolase from thermus thermophilus HB8 | | Descriptor: | 1-HYDROXY-PENTANE-3,4-DIOL-5-PHOSPHATE, Aldolase protein | | Authors: | Lokanath, N.K, Miyano, M, Yokoyama, S, Kuramitsu, S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-03-28 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of aldolase from Thermus thermophilus HB8 showing the contribution of oligomeric state to thermostability.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
8IYH
 
 | | Structure of MK6892-GPR109A-G-protein complex | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-04-04 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution.
Nat Commun, 15, 2024
|
|
8IYW
 
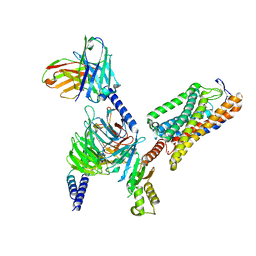 | | Structure of GSK256073-GPR109A-G-protein complex | | Descriptor: | 8-chloranyl-3-pentyl-7H-purine-2,6-dione, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution.
Nat Commun, 15, 2024
|
|
8JHN
 
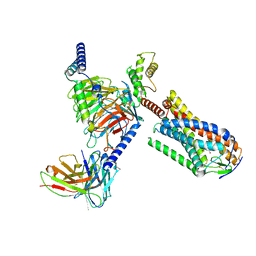 | | Structure of MMF-GPR109A-G protein complex | | Descriptor: | (E)-4-methoxy-4-oxidanylidene-but-2-enoic acid, G protein subunit alpha o1,Guanine nucleotide-binding protein G(o) subunit alpha, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-05-24 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution.
Nat Commun, 15, 2024
|
|
