5Y03
 
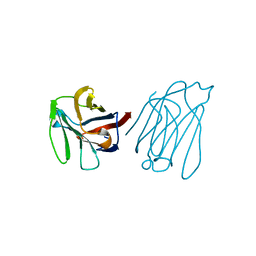 | | Galectin-13/Placental Protein 13 variant R53H crystal structure | | Descriptor: | Galactoside-binding soluble lectin 13 | | Authors: | Wang, Y, Su, J. | | Deposit date: | 2017-07-14 | | Release date: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Galectin-13, a different prototype galectin, does not bind beta-galacto-sides and forms dimers via intermolecular disulfide bridges between Cys-136 and Cys-138.
Sci Rep, 8, 2018
|
|
5XXH
 
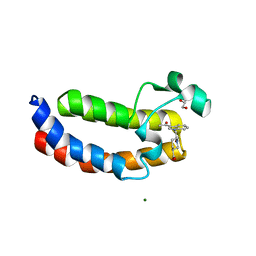 | | Crystal Structure Analysis of the CBP | | Descriptor: | (3S)-1-[2-(3-ethanoylindol-1-yl)ethanoyl]piperidine-3-carboxylic acid, 1,2-ETHANEDIOL, CREB-binding protein, ... | | Authors: | Xiang, Q, Zhang, Y, Wang, C, Song, M. | | Deposit date: | 2017-07-04 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and optimization of 1-(1H-indol-1-yl)ethanone derivatives as CBP/EP300 bromodomain inhibitors for the treatment of castration-resistant prostate cancer.
Eur J Med Chem, 147, 2018
|
|
7DAJ
 
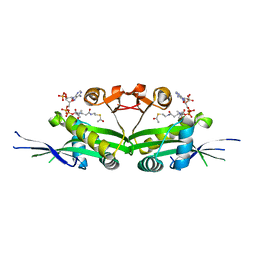 | | The crystal structure of serotonin N-acetyltransferase in complex with acetyl-CoA from Oryza Sativa | | Descriptor: | ACETYL COENZYME *A, Serotonin N-acetyltransferase 1, chloroplastic | | Authors: | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | Deposit date: | 2020-10-16 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DAI
 
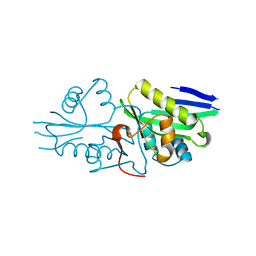 | | The crystal structure of a serotonin N-acetyltransferase from Oryza Sativa | | Descriptor: | Serotonin N-acetyltransferase 1, chloroplastic | | Authors: | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | Deposit date: | 2020-10-16 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DAL
 
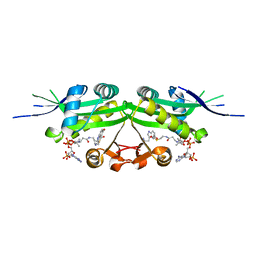 | | The crystal structure of a serotonin N-acetyltransferase in complex with serotonin and acetyl-CoA from Oryza Sativa | | Descriptor: | ACETYL COENZYME *A, SEROTONIN, Serotonin N-acetyltransferase 1, ... | | Authors: | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | Deposit date: | 2020-10-16 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DAK
 
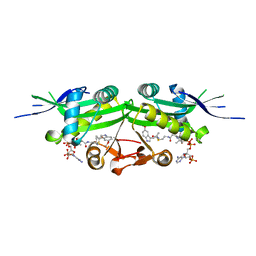 | | The crystal structure of a serotonin N-acetyltransferase in complex with 5-Methoxytryptamine and acetyl-CoA from Oryza Sativa | | Descriptor: | 2-(5-methoxy-1H-indol-3-yl)ethanamine, ACETYL COENZYME *A, Serotonin N-acetyltransferase 1, ... | | Authors: | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | Deposit date: | 2020-10-16 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5XRG
 
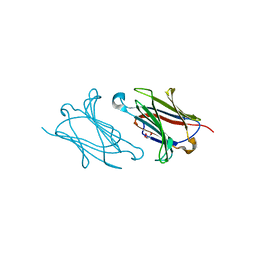 | | Galectin-10/Charcot-Leyden crystal protein crystal structure | | Descriptor: | GLYCEROL, Galectin-10 | | Authors: | Su, J. | | Deposit date: | 2017-06-08 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Galectin-10: a new structural type of prototype galectin dimer and effects on saccharide ligand binding.
Glycobiology, 28, 2018
|
|
5XRK
 
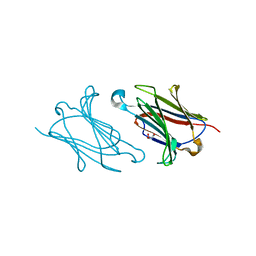 | |
5XRP
 
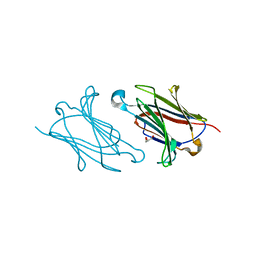 | | Galectin-10/Charcot-Leyden crystal protein variant Q75A | | Descriptor: | GLYCEROL, Galectin-10 | | Authors: | Su, J. | | Deposit date: | 2017-06-08 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Galectin-10: a new structural type of prototype galectin dimer and effects on saccharide ligand binding.
Glycobiology, 28, 2018
|
|
5XRO
 
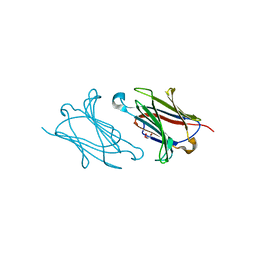 | |
5XRI
 
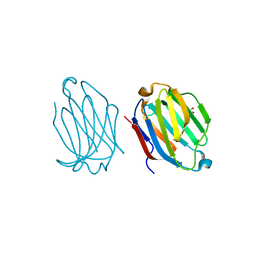 | |
5XRM
 
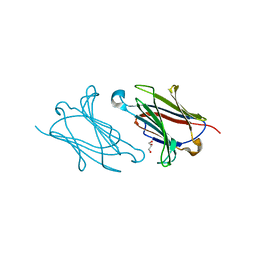 | |
5XRH
 
 | | Galectin-10/Charcot-Leyden crystal protein crystal structure | | Descriptor: | Galectin-10 | | Authors: | Su, J. | | Deposit date: | 2017-06-08 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Galectin-10: a new structural type of prototype galectin dimer and effects on saccharide ligand binding.
Glycobiology, 28, 2018
|
|
5XRN
 
 | |
5YT4
 
 | | Galectin-10 variant H53A soaked in glycerol for 5 minutes | | Descriptor: | GLYCEROL, Galectin-10 | | Authors: | Su, J. | | Deposit date: | 2017-11-16 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Galectin-10: a new structural type of prototype galectin dimer and effects on saccharide ligand binding.
Glycobiology, 28, 2018
|
|
5XRL
 
 | |
5XRJ
 
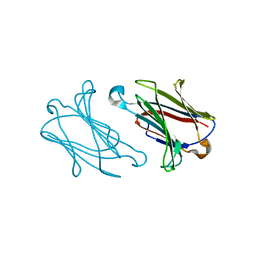 | |
7COM
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Boceprevir (space group P212121) | | Descriptor: | 3C-like proteinase, boceprevir (bound form) | | Authors: | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Liu, J.M, Zhou, Y.L, Chen, P, Yang, S.Y, Lei, J. | | Deposit date: | 2020-08-04 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
5YHQ
 
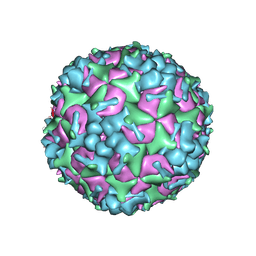 | | Cryo-EM Structure of CVA6 VLP | | Descriptor: | Capsid protein VP1, Capsid protein VP3, capsid protein VP0 | | Authors: | Chen, J, Zhang, C, Huang, Z, Cong, Y. | | Deposit date: | 2017-09-29 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A 3.0-Angstrom Resolution Cryo-Electron Microscopy Structure and Antigenic Sites of Coxsackievirus A6-Like Particles.
J. Virol., 92, 2018
|
|
7F58
 
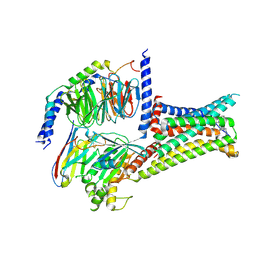 | | Cryo-EM structure of THIQ-MC4R-Gs_Nb35 complex | | Descriptor: | (3R)-N-[(2R)-3-(4-chlorophenyl)-1-[4-cyclohexyl-4-(1,2,4-triazol-1-ylmethyl)piperidin-1-yl]-1-oxidanylidene-propan-2-yl]-1,2,3,4-tetrahydroisoquinoline-3-carboxamide, CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, H, Chen, L, Mao, C, Shen, Q, Yang, D, Shen, D, Qin, J. | | Deposit date: | 2021-06-21 | | Release date: | 2021-11-03 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into ligand recognition and activation of the melanocortin-4 receptor.
Cell Res., 31, 2021
|
|
7F54
 
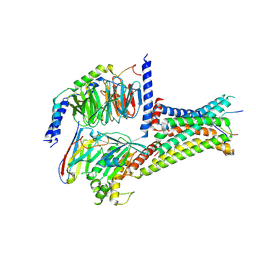 | | Cryo-EM structure of afamelanotide-MC4R-Gs_Nb35 complex | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, H, Chen, L, Mao, C, Shen, Q, Yang, D, Shen, D, Qin, J. | | Deposit date: | 2021-06-21 | | Release date: | 2021-11-03 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into ligand recognition and activation of the melanocortin-4 receptor.
Cell Res., 31, 2021
|
|
7F55
 
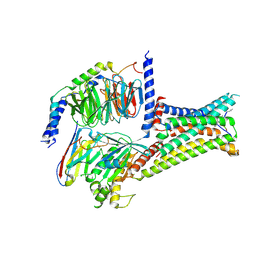 | | Cryo-EM structure of bremelanotide-MC4R-Gs_Nb35 complex | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, H, Chen, L, Mao, C, Shen, Q, Yang, D, Shen, D, Qin, J. | | Deposit date: | 2021-06-21 | | Release date: | 2021-11-03 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into ligand recognition and activation of the melanocortin-4 receptor.
Cell Res., 31, 2021
|
|
7F53
 
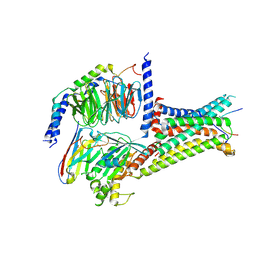 | | Cryo-EM structure of a-MSH-MC4R-Gs_Nb35 complex | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, H, Chen, L, Mao, C, Shen, Q, Yang, D, Shen, D, Qin, J. | | Deposit date: | 2021-06-21 | | Release date: | 2021-11-03 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into ligand recognition and activation of the melanocortin-4 receptor.
Cell Res., 31, 2021
|
|
8HGW
 
 | | Crystal structure of MehpH in complex with MBP | | Descriptor: | 1-BUTANOL, Monoalkyl phthalate hydrolase, PHTHALIC ACID | | Authors: | Zhang, Z.M, Wang, Y.J, Chen, Y.B. | | Deposit date: | 2022-11-15 | | Release date: | 2023-03-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.80001163 Å) | | Cite: | Molecular insights into the catalytic mechanism of plasticizer degradation by a monoalkyl phthalate hydrolase.
Commun Chem, 6, 2023
|
|
8HGV
 
 | |
