5YLE
 
 | | MCR-1 complex with ethanolamine (ETA) | | Descriptor: | ETHANOLAMINE, Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION | | Authors: | Wei, P.C, Song, G.J, Shi, M.Y, Zhou, Y.F, Liu, Y, Lei, J, Chen, P, Yin, L. | | Deposit date: | 2017-10-17 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate analog interaction with MCR-1 offers insight into the rising threat of the plasmid-mediated transferable colistin resistance.
FASEB J., 32, 2018
|
|
2J6W
 
 | | R164N mutant of the RUNX1 Runt domain | | Descriptor: | CHLORIDE ION, RUNT-RELATED TRANSCRIPTION FACTOR 1 | | Authors: | Grembecka, J, Zhe, L, Lukasik, S.M, Liu, Y, Bielnicka, I, Bushweller, J.H, Speck, N.A. | | Deposit date: | 2006-10-04 | | Release date: | 2007-10-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Mutation in the S-Switch Region of the Runt Domain Alters the Dynamics of an Allosteric Network Responsible for Cbfbeta Regulation.
J.Mol.Biol., 364, 2006
|
|
7X3M
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07045 | | Descriptor: | (2~{R})-2-[4-[3,5-bis(chloranyl)phenyl]-3-(trifluoromethyl)phenyl]butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
7X3L
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07044 | | Descriptor: | (2~{R})-2-[4-(3-fluoranyl-4-methyl-phenyl)-3-(trifluoromethyl)phenyl]butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
2JVC
 
 | |
2N3J
 
 | | Solution Structure of the alpha-crystallin domain from the redox-sensitive chaperone, HSPB1 | | Descriptor: | Heat shock protein beta-1 | | Authors: | Rajagopal, P, Liu, Y, Shi, L, Klevit, R.E. | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the alpha-crystallin domain from the redox-sensitive chaperone, HSPB1.
J.Biomol.Nmr, 63, 2015
|
|
2OIG
 
 | | Crystal structure of RS21-C6 core segment and dm5CTP complex | | Descriptor: | 2'-DEOXY-5-METHYLCYTIDINE 5'-(TETRAHYDROGEN TRIPHOSPHATE), RS21-C6 | | Authors: | Wu, B, Liu, Y, Zhao, Q, Liao, S, Zhang, J, Bartlam, M, Chen, W, Rao, Z. | | Deposit date: | 2007-01-11 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of RS21-C6, Involved in Nucleoside Triphosphate Pyrophosphohydrolysis
J.Mol.Biol., 367, 2007
|
|
2OIE
 
 | | Crystal structure of RS21-C6 core segment RSCUT | | Descriptor: | RS21-C6, SULFATE ION | | Authors: | Wu, B, Liu, Y, Zhao, Q, Liao, S, Zhang, J, Bartlam, M, Chen, W, Rao, Z. | | Deposit date: | 2007-01-10 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of RS21-C6, Involved in Nucleoside Triphosphate Pyrophosphohydrolysis
J.Mol.Biol., 367, 2007
|
|
2O0F
 
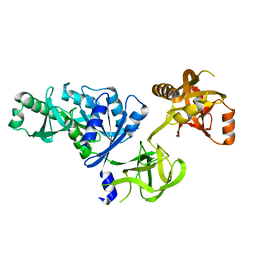 | | Docking of the modified RF3 X-ray structure into cryo-EM map of E.coli 70S ribosome bound with RF3 | | Descriptor: | Peptide chain release factor 3 | | Authors: | Gao, H, Zhou, Z, Rawat, U, Huang, C, Bouakaz, L, Wang, C, Liu, Y, Zavialov, A, Gursky, R, Sanyal, S, Ehrenberg, M, Frank, J, Song, H. | | Deposit date: | 2006-11-27 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (15.5 Å) | | Cite: | RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors
Cell(Cambridge,Mass.), 129, 2007
|
|
2LQ9
 
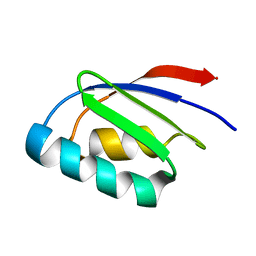 | |
4LYP
 
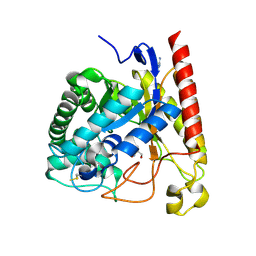 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Exo-beta-1,4-mannosidase, GUANIDINE | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8H69
 
 | | Cryo-EM structure of influenza RNA polymerase | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*UP*AP*AP*AP*CP*UP*CP*CP*UP*GP*CP*UP*UP*UP*UP*GP*CP*U)-3'), ... | | Authors: | Li, H, Wu, Y, Liang, H, Liu, Y. | | Deposit date: | 2022-10-16 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | An intermediate state allows influenza polymerase to switch smoothly between transcription and replication cycles.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4NRR
 
 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase (E202A mutant) from Rhizomucor miehei in complex with mannosyl-fructose | | Descriptor: | Exo-beta-1,4-mannosidase, beta-D-mannopyranose-(1-4)-beta-D-fructofuranose | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-11-27 | | Release date: | 2014-11-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8WY0
 
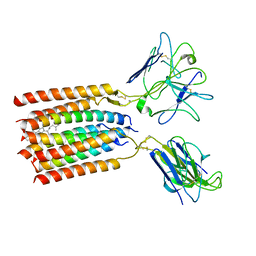 | | T cell receptor delta 2 gamma 9 with F283A, F290A, and F291A | | Descriptor: | CHOLESTEROL, Signal peptide,flag tag,T cell receptor delta variable 2,T cell receptor delta constant, Signal peptide,flag tag,T cell receptor gamma variable 9,T cell receptor gamma constant 1, ... | | Authors: | Xin, W, Huang, B, Chi, X, Liu, Y, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | Deposit date: | 2023-10-30 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
8WYI
 
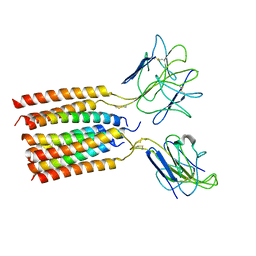 | | T cell receptor delta 2 gamma 9 with TCRD TM domain chimera of TRAC | | Descriptor: | Signal peptide,flag tag,T cell receptor delta variable 2,T cell receptor delta constant,T cell receptor alpha chain constant,T cell receptor delta variable 2,T cell receptor delta constant,T cell receptor alpha chain constant, Signal peptide,flag tag,T cell receptor gamma variable 9,T cell receptor gamma constant 1,T cell receptor gamma variable 9,T cell receptor gamma constant 1, T-cell surface glycoprotein CD3 delta chain, ... | | Authors: | Xin, W, Huang, B, Chi, X, Liu, Y, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
4NRS
 
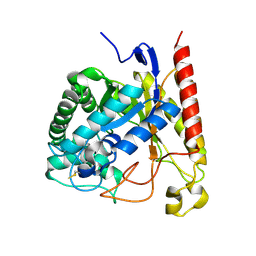 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase (E202A mutant) from Rhizomucor miehei in complex with mannobiose | | Descriptor: | Exo-beta-1,4-mannosidase, beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-11-27 | | Release date: | 2014-11-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8WX8
 
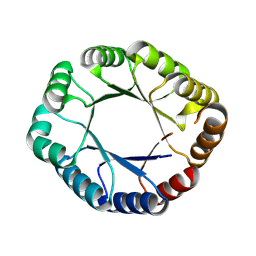 | |
8WWC
 
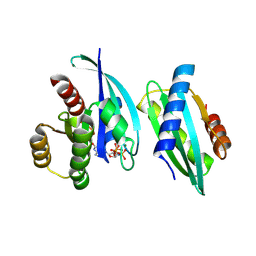 | |
3UA3
 
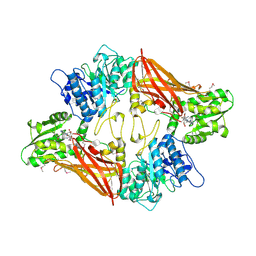 | | Crystal Structure of Protein Arginine Methyltransferase PRMT5 in complex with SAH | | Descriptor: | Protein arginine N-methyltransferase 5, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Sun, L, Wang, M, Lv, Z, Yang, N, Liu, Y, Bao, S, Gong, W, Xu, R.M. | | Deposit date: | 2011-10-20 | | Release date: | 2011-12-14 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into protein arginine symmetric dimethylation by PRMT5
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3UA4
 
 | | Crystal Structure of Protein Arginine Methyltransferase PRMT5 | | Descriptor: | GLYCEROL, Protein arginine N-methyltransferase 5 | | Authors: | Sun, L, Wang, M, Lv, Z, Yang, N, Liu, Y, Bao, S, Gong, W, Xu, R.M. | | Deposit date: | 2011-10-21 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Structural insights into protein arginine symmetric dimethylation by PRMT5
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7DPD
 
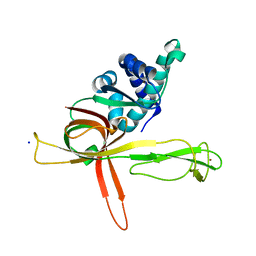 | | Human MCM9 N-terminal domain | | Descriptor: | DNA helicase MCM9, SODIUM ION, ZINC ION | | Authors: | Li, J, Liu, L, Liu, Y. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural study of the N-terminal domain of human MCM8/9 complex.
Structure, 29, 2021
|
|
7DP3
 
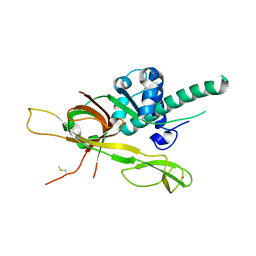 | | Human MCM8 N-terminal domain | | Descriptor: | DNA helicase MCM8, ZINC ION | | Authors: | Li, J, Liu, L, Liu, Y. | | Deposit date: | 2020-12-17 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural study of the N-terminal domain of human MCM8/9 complex.
Structure, 29, 2021
|
|
5ZUE
 
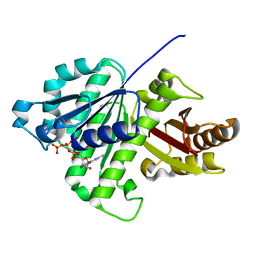 | | GTP-bound, double-stranded, curved FtsZ protofilament structure | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Guan, F, Yu, J, Yu, J, Liu, Y, Li, Y, Feng, X.H, Huang, K.C, Chang, Z, Ye, S. | | Deposit date: | 2018-05-07 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Lateral interactions between protofilaments of the bacterial tubulin homolog FtsZ are essential for cell division
Elife, 7, 2018
|
|
6B0V
 
 | | Crystal Structure of small molecule ARS-107 covalently bound to K-Ras G12C | | Descriptor: | 1-[3-(4-{[(4,5-dichloro-2-hydroxyphenyl)amino]acetyl}piperazin-1-yl)azetidin-1-yl]propan-1-one, CALCIUM ION, GTPase KRas, ... | | Authors: | Hansen, R, Peters, U, Babbar, A, Chen, Y, Feng, J, Janes, M.R, Li, L.-S, Ren, P, Liu, Y, Zarrinkar, P.P. | | Deposit date: | 2017-09-15 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | The reactivity-driven biochemical mechanism of covalent KRASG12Cinhibitors.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6B0Y
 
 | | Crystal Structure of small molecule ARS-917 covalently bound to K-Ras G12C | | Descriptor: | 1-{4-[6-chloro-7-(2-fluorophenyl)quinazolin-4-yl]piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | Authors: | Hansen, R, Peters, U, Babbar, A, Chen, Y, Feng, J, Janes, M.R, Li, L.-S, Ren, P, Liu, Y, Zarrinkar, P.P. | | Deposit date: | 2017-09-15 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The reactivity-driven biochemical mechanism of covalent KRASG12Cinhibitors.
Nat. Struct. Mol. Biol., 25, 2018
|
|
