8FLK
 
 | | Cryo-EM structure of STING oligomer bound to cGAMP and NVS-STG2 | | Descriptor: | 4-({[4-(2-tert-butyl-5,5-dimethyl-1,3-dioxan-2-yl)phenyl]methyl}amino)-3-methoxybenzoic acid, Stimulator of interferon genes protein, cGAMP | | Authors: | Li, J, Canham, S.M, Zhang, X, Bai, X, Feng, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-01 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Activation of human STING by a molecular glue-like compound.
Nat.Chem.Biol., 20, 2024
|
|
2DHR
 
 | | Whole cytosolic region of ATP-dependent metalloprotease FtsH (G399L) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FtsH | | Authors: | Suno, R, Niwa, H, Tsuchiya, D, Zhang, X, Yoshida, M, Morikawa, K. | | Deposit date: | 2006-03-24 | | Release date: | 2006-06-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of the Whole Cytosolic Region of ATP-Dependent Protease FtsH
Mol.Cell, 22, 2006
|
|
2DI4
 
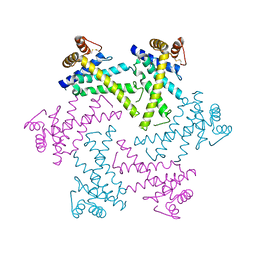 | | Crystal structure of the FtsH protease domain | | Descriptor: | Cell division protein ftsH homolog, MERCURY (II) ION | | Authors: | Suno, R, Niwa, H, Tsuchiya, D, Zhang, X, Yoshida, M, Morikawa, K. | | Deposit date: | 2006-03-28 | | Release date: | 2006-06-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure of the Whole Cytosolic Region of ATP-Dependent Protease FtsH
Mol.Cell, 22, 2006
|
|
6M3W
 
 | | Post-fusion structure of SARS-CoV spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Fan, X, Cao, D, Zhang, X. | | Deposit date: | 2020-03-04 | | Release date: | 2020-06-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM analysis of the post-fusion structure of the SARS-CoV spike glycoprotein.
Nat Commun, 11, 2020
|
|
8YZD
 
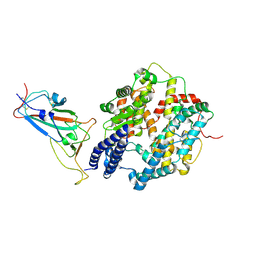 | | Structure of JN.1 RBD protein in complex with ACE2. | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin,fusion protein | | Authors: | Wang, Y.J, Zhang, X, Sun, L. | | Deposit date: | 2024-04-06 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Lineage-specific pathogenicity, immune evasion, and virological features of SARS-CoV-2 BA.2.86/JN.1 and EG.5.1/HK.3.
Nat Commun, 15, 2024
|
|
8YZE
 
 | | The JN.1 spike protein (S) in complex with ACE2. | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin,Expression Tag | | Authors: | Wang, Y.J, Zhang, X, Sun, L. | | Deposit date: | 2024-04-06 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Lineage-specific pathogenicity, immune evasion, and virological features of SARS-CoV-2 BA.2.86/JN.1 and EG.5.1/HK.3.
Nat Commun, 15, 2024
|
|
8YZB
 
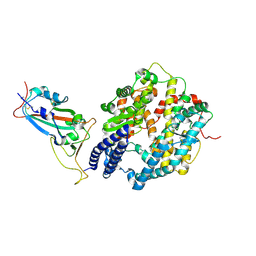 | | BA.2.86 RBD protein in complex with ACE2. | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin,fusion protein | | Authors: | Wang, Y.J, Zhang, X, Sun, L. | | Deposit date: | 2024-04-06 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Lineage-specific pathogenicity, immune evasion, and virological features of SARS-CoV-2 BA.2.86/JN.1 and EG.5.1/HK.3.
Nat Commun, 15, 2024
|
|
2C9C
 
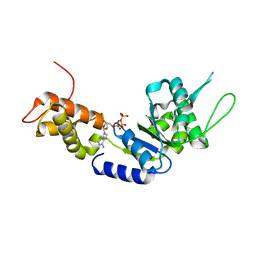 | | Structural basis of the nucleotide driven conformational changes in the AAA domain of transcription activator PspF | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PSP OPERON TRANSCRIPTIONAL ACTIVATOR | | Authors: | Rappas, M, Schumacher, J, Niwa, H, Buck, M, Zhang, X. | | Deposit date: | 2005-12-09 | | Release date: | 2006-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of the Nucleotide Driven Conformational Changes in the Aaa(+) Domain of Transcription Activator Pspf.
J.Mol.Biol., 357, 2006
|
|
2C98
 
 | | Structural basis of the nucleotide driven conformational changes in the AAA domain of transcription activator PspF | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PSP OPERON TRANSCRIPTIONAL ACTIVATOR | | Authors: | Rappas, M, Schumacher, J, Niwa, H, Buck, M, Zhang, X. | | Deposit date: | 2005-12-09 | | Release date: | 2006-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of the Nucleotide Driven Conformational Changes in the Aaa(+) Domain of Transcription Activator Pspf.
J.Mol.Biol., 357, 2006
|
|
2C99
 
 | | Structural basis of the nucleotide driven conformational changes in the AAA domain of transcription activator PspF | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PSP OPERON TRANSCRIPTIONAL ACTIVATOR | | Authors: | Rappas, M, Schumacher, J, Niwa, H, Buck, M, Zhang, X. | | Deposit date: | 2005-12-09 | | Release date: | 2006-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of the Nucleotide Driven Conformational Changes in the Aaa(+) Domain of Transcription Activator Pspf.
J.Mol.Biol., 357, 2006
|
|
2C96
 
 | | Structural basis of the nucleotide driven conformational changes in the AAA domain of transcription activator PspF | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PSP OPERON TRANSCRIPTIONAL ACTIVATOR | | Authors: | Rappas, M, Schumacher, J, Niwa, H, Buck, M, Zhang, X. | | Deposit date: | 2005-12-09 | | Release date: | 2006-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of the Nucleotide Driven Conformational Changes in the Aaa(+) Domain of Transcription Activator Pspf.
J.Mol.Biol., 357, 2006
|
|
6IYA
 
 | |
2H3B
 
 | | Crystal Structure of Mouse Nicotinamide Phosphoribosyltransferase/Visfatin/Pre-B Cell Colony Enhancing Factor 1 | | Descriptor: | Nicotinamide phosphoribosyltransferase, SULFATE ION | | Authors: | Wang, T, Zhang, X, Bheda, P, Revollo, J.R, Imai, S.I, Wolberger, C. | | Deposit date: | 2006-05-22 | | Release date: | 2006-06-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Nampt/PBEF/visfatin, a mammalian NAD(+) biosynthetic enzyme.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2GZY
 
 | |
2GIC
 
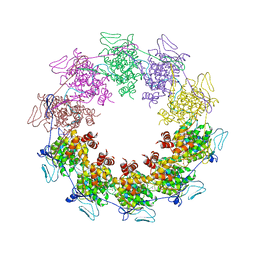 | | Crystal Structure of a vesicular stomatitis virus nucleocapsid-RNA complex | | Descriptor: | 45-MER, Nucleocapsid protein, URANYL (VI) ION | | Authors: | Green, T.J, Zhang, X, Wertz, G.W, Luo, M. | | Deposit date: | 2006-03-28 | | Release date: | 2006-08-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structure of the vesicular stomatitis virus nucleoprotein-RNA complex unveils how the RNA is sequestered
Science, 313, 2006
|
|
2GZZ
 
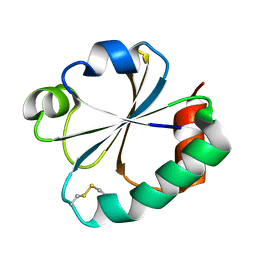 | |
2H3D
 
 | | Crystal Structure of Mouse Nicotinamide Phosphoribosyltransferase/Visfatin/Pre-B Cell Colony Enhancing Factor in Complex with Nicotinamide Mononuleotide | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Nicotinamide phosphoribosyltransferase | | Authors: | Wang, T, Zhang, X, Bheda, P, Revollo, J.R, Imai, S.I, Wolberger, C. | | Deposit date: | 2006-05-22 | | Release date: | 2006-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Nampt/PBEF/visfatin, a mammalian NAD(+) biosynthetic enzyme.
Nat.Struct.Mol.Biol., 13, 2006
|
|
7Y1A
 
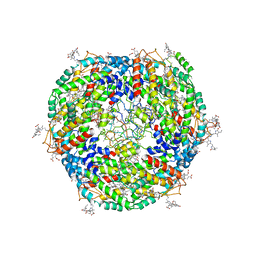 | | Lateral hexamer | | Descriptor: | B-phycoerythrin beta chain, LRH, PHYCOERYTHROBILIN, ... | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | Deposit date: | 2022-06-07 | | Release date: | 2023-01-18 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
2L37
 
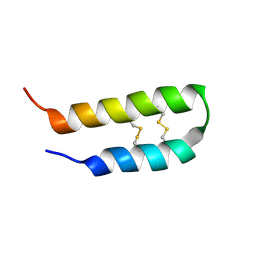 | | 3D solution structure of arginine/glutamate-rich polypeptide Luffin P1 from the seeds of sponge gourd (Luffa cylindrical) | | Descriptor: | Ribosome-inactivating protein luffin P1 | | Authors: | Ng, Y.M, Yang, Y, Sze, K.H, Zhang, X, Zheng, Y.T, Shaw, P.C. | | Deposit date: | 2010-09-08 | | Release date: | 2011-01-19 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural characterization and anti-HIV-1 activities of arginine/glutamate-rich polypeptide Luffin P1 from the seeds of sponge gourd (Luffa cylindrical).
J.Struct.Biol., 2010
|
|
2LUH
 
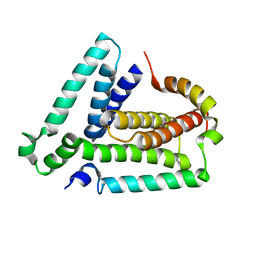 | | NMR structure of the Vta1-Vps60 complex | | Descriptor: | Vacuolar protein sorting-associated protein VTA1, Vacuolar protein-sorting-associated protein 60 | | Authors: | Yang, Z, Vild, C, Ju, J, Zhang, X, Liu, J, Shen, J, Zhao, B, Lan, W, Gong, F, Liu, M, Cao, C, Xu, Z. | | Deposit date: | 2012-06-13 | | Release date: | 2012-11-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Molecular Recognition between ESCRT-III-like Protein Vps60 and AAA-ATPase Regulator Vta1 in the Multivesicular Body Pathway.
J.Biol.Chem., 287, 2012
|
|
7Y4L
 
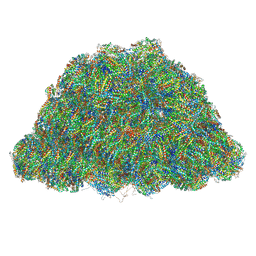 | | PBS of PBS-PSII-PSI-LHCs from Porphyridium purpureum. | | Descriptor: | Allophycocyanin alpha subunit, Allophycocyanin beta 18 subunit, Allophycocyanin beta subunit, ... | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | Deposit date: | 2022-06-15 | | Release date: | 2023-01-18 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
7Y5E
 
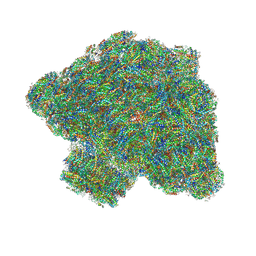 | | In situ single-PBS-PSII-PSI-LHCs megacomplex. | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (2S)-2,3-dihydroxypropyl octadecanoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sui, S.F. | | Deposit date: | 2022-06-17 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
7Y7A
 
 | | In situ double-PBS-PSII-PSI-LHCs megacomplex from Porphyridium purpureum. | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (2S)-2,3-dihydroxypropyl octadecanoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | Deposit date: | 2022-06-22 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
6MS7
 
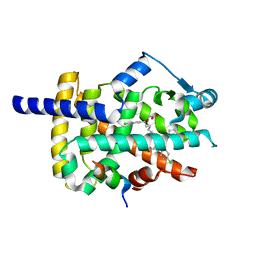 | | Peroxisome proliferator-activated receptor gamma ligand binding domain in complex with a novel selective PPAR-gamma modulator VSP-77 | | Descriptor: | PGC1 LXXLL motif, Peroxisome proliferator-activated receptor gamma, {[(1S)-1-(4-chlorophenyl)octyl]oxy}acetic acid | | Authors: | Yi, W, Jiang, H, Zhou, X.E, Shi, J, Zhao, G, Zhang, X, Sun, Y, Suino-Powell, K, Li, J, Li, J, Melcher, K, Xu, H.E. | | Deposit date: | 2018-10-16 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Identification and structural insight of an effective PPAR gamma modulator with improved therapeutic index for anti-diabetic drug discovery.
Chem Sci, 11, 2020
|
|
2K2V
 
 | |
