6NHR
 
 | |
6NSA
 
 | |
6NSG
 
 | |
6N35
 
 | | Anti-HIV-1 Fab 2G12 + Man1-2 re-refinement | | Descriptor: | BENZOIC ACID, Fab 2G12 heavy chain, Fab 2G12 light chain, ... | | Authors: | Calarese, D.A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2018-11-14 | | Release date: | 2018-11-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Antibody domain exchange is an immunological solution to carbohydrate cluster recognition.
Science, 300, 2003
|
|
6NSC
 
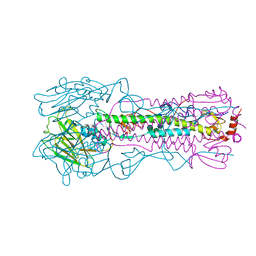 | | Crystal structure of the A/Brisbane/10/2007 (H3N2) influenza virus hemagglutinin G186V/L194P mutant apo form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, N.C, Wilson, I.A. | | Deposit date: | 2019-01-24 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Preventing an Antigenically Disruptive Mutation in Egg-Based H3N2 Seasonal Influenza Vaccines by Mutational Incompatibility.
Cell Host Microbe, 25, 2019
|
|
6O3U
 
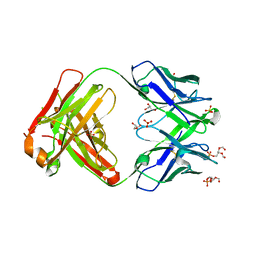 | |
6NS9
 
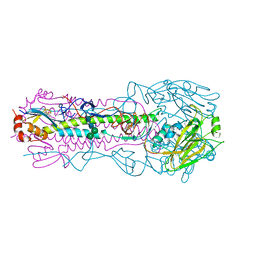 | | Crystal structure of the IVR-165 (H3N2) influenza virus hemagglutinin apo form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Wu, N.C, Wilson, I.A. | | Deposit date: | 2019-01-24 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Preventing an Antigenically Disruptive Mutation in Egg-Based H3N2 Seasonal Influenza Vaccines by Mutational Incompatibility.
Cell Host Microbe, 25, 2019
|
|
6NSF
 
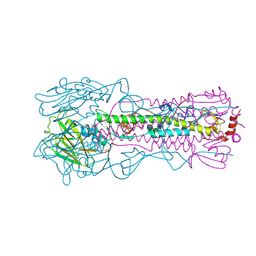 | |
6N32
 
 | | Anti-HIV-1 Fab 2G12 re-refinement | | Descriptor: | Fab 2G12 heavy chain, Fab 2G12 light chain, SULFATE ION | | Authors: | Calarese, D.A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2018-11-14 | | Release date: | 2018-11-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antibody domain exchange is an immunological solution to carbohydrate cluster recognition.
Science, 300, 2003
|
|
6NHQ
 
 | |
6O3D
 
 | |
6O3J
 
 | | Crystal structure of the Fab fragment of the human HIV-1 neutralizing antibody PGZL1 in complex with its MPER peptide epitope (region 671-683 of HIV-1 gp41) and phosphatidic acid (06:0 PA) | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, (4S)-2-METHYL-2,4-PENTANEDIOL, MPER peptide, ... | | Authors: | Irimia, A, Wilson, I.A. | | Deposit date: | 2019-02-26 | | Release date: | 2019-12-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.416 Å) | | Cite: | An MPER antibody neutralizes HIV-1 using germline features shared among donors.
Nat Commun, 10, 2019
|
|
6O30
 
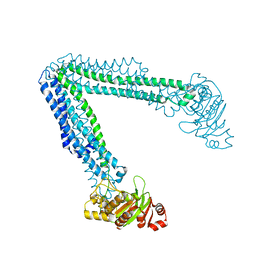 | | Lipid A transporter MsbA from Salmonella typhimurium | | Descriptor: | Lipid A export ATP-binding/permease protein MsbA | | Authors: | Padayatti, P.S, Zhang, Q, Wilson, I.A, Lee, S.C, Stanfield, R.L. | | Deposit date: | 2019-02-25 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.47 Å) | | Cite: | Structural Insights into the Lipid A Transport Pathway in MsbA.
Structure, 27, 2019
|
|
6O3L
 
 | |
6O42
 
 | |
6O3K
 
 | |
1XZ0
 
 | | Crystal structure of CD1a in complex with a synthetic mycobactin lipopeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(HYDROXY-HEXADECANOYL-AMINO)-2-{[(4S)-2-(2-HYDROXY-PHENYL)-4,5-DIHYDRO-OXAZOLE-4-CARBONYL]-AMINO}-HEXANOIC ACID 2-[(3S)-1-(TERT-BUTYL-DIPHENYL-SILANYLOXY)-2-OXO-AZEPAN-3-YLCARBAMOYL]-(1S)-1-METHYL-ETHYL ESTER, Beta-2-microglobulin, ... | | Authors: | Zajonc, D.M, Crispin, M.D, Bowden, T.A, Young, D.C, Cheng, T.Y, Hu, J, Costello, C.E, Miller, M.J, Moody, D.B, Wilson, I.A. | | Deposit date: | 2004-11-11 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular Mechanism of Lipopeptide Presentation by CD1a.
Immunity, 22, 2005
|
|
6OO0
 
 | | Crystal structure of bovine Fab NC-Cow1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, NC-Cow1 heavy chain, NC-Cow1 light chain | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2019-04-22 | | Release date: | 2020-04-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of broad HIV neutralization by a vaccine-induced cow antibody.
Sci Adv, 6, 2020
|
|
1Y18
 
 | | Fab fragment of catalytic elimination antibody 34E4 E(H50)D mutant in complex with hapten | | Descriptor: | 2-AMINO-5,6-DIMETHYL-BENZIMIDAZOLE-1-PENTANOIC ACID, CHLORIDE ION, Catalytic antibody 34E4 heavy chain, ... | | Authors: | Debler, E.W, Ito, S, Heine, A, Wilson, I.A. | | Deposit date: | 2004-11-17 | | Release date: | 2005-04-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural origins of efficient proton abstraction from carbon by a catalytic antibody
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1Y0L
 
 | | Catalytic elimination antibody 34E4 in complex with hapten | | Descriptor: | 2-AMINO-5,6-DIMETHYL-BENZIMIDAZOLE-1-PENTANOIC ACID, CHLORIDE ION, Catalytic Antibody Fab 34E4 Heavy chain, ... | | Authors: | Debler, E.W, Ito, S, Heine, A, Wilson, I.A. | | Deposit date: | 2004-11-15 | | Release date: | 2005-04-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural origins of efficient proton abstraction from carbon by a catalytic antibody
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1A3L
 
 | |
1AI1
 
 | | HIV-1 V3 LOOP MIMIC | | Descriptor: | AIB142, IGG1-KAPPA 59.1 FAB (HEAVY CHAIN), IGG1-KAPPA 59.1 FAB (LIGHT CHAIN) | | Authors: | Ghiara, J.B, Wilson, I.A. | | Deposit date: | 1996-11-06 | | Release date: | 1997-05-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of a constrained peptide mimic of the HIV-1 V3 loop neutralization site.
J.Mol.Biol., 266, 1997
|
|
1AHW
 
 | | A COMPLEX OF EXTRACELLULAR DOMAIN OF TISSUE FACTOR WITH AN INHIBITORY FAB (5G9) | | Descriptor: | IMMUNOGLOBULIN FAB 5G9 (HEAVY CHAIN), IMMUNOGLOBULIN FAB 5G9 (LIGHT CHAIN), TISSUE FACTOR | | Authors: | Huang, M, Syed, R, Stura, E.A, Stone, M.J, Stefanko, R.S, Ruf, W, Edgington, T.S, Wilson, I.A. | | Deposit date: | 1997-04-10 | | Release date: | 1998-02-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The mechanism of an inhibitory antibody on TF-initiated blood coagulation revealed by the crystal structures of human tissue factor, Fab 5G9 and TF.5G9 complex.
J.Mol.Biol., 275, 1998
|
|
1ACY
 
 | |
1AXT
 
 | |
