9OA7
 
 | | GNAT family acetyltransferase EryM SeMet | | Descriptor: | GLYCEROL, Lysine N-acyltransferase MbtK, MAGNESIUM ION, ... | | Authors: | Li, Y, Smith, J. | | Deposit date: | 2025-04-19 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Redefining the role of the EryM acetyltransferase in natural product biosynthetic pathways.
Structure, 2025
|
|
9NNQ
 
 | | GNAT family acetyltransferase EryM | | Descriptor: | GLYCEROL, Lysine N-acyltransferase MbtK, MAGNESIUM ION, ... | | Authors: | Li, Y, Smith, J. | | Deposit date: | 2025-03-06 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Redefining the role of the EryM acetyltransferase in natural product biosynthetic pathways.
Structure, 2025
|
|
9LSL
 
 | | The crystal structure of PDE5A with L1 | | Descriptor: | (13~{S},15~{R})-15-(3-chloranyl-4-methoxy-phenyl)-12-ethanoyl-8,12,16-triazatetracyclo[7.7.1.0^{2,7}.0^{13,17}]heptadeca-1(17),2,4,6,8-pentaene-4-carbonitrile, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wu, D, Huang, Y.-Y, Luo, H.-B. | | Deposit date: | 2025-02-04 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Plasma metabolites-based drug design: Discovery of novel and highly selective phosphodiesterase 5 inhibitors
Chin.Chem.Lett., 2025
|
|
5GZR
 
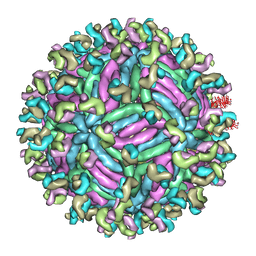 | | Zika virus E protein complexed with a neutralizing antibody Z23-Fab | | Descriptor: | Z23 Fab heavy chain, Z23 Fab light chain, structural protein E, ... | | Authors: | Gao, G.G, Shi, Y, Peng, R, Liu, S. | | Deposit date: | 2016-10-01 | | Release date: | 2016-11-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
8HLC
 
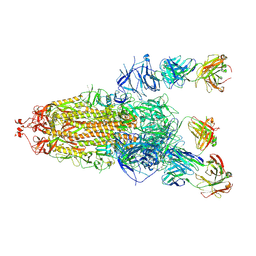 | | S protein of SARS-CoV-2 in complex with 3711 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Y.Y, Guo, Y.Y, Zhou, Q. | | Deposit date: | 2022-11-29 | | Release date: | 2024-06-05 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Defining the features and structure of neutralizing antibody targeting the silent face of the SARS-CoV-2 spike N-terminal domain.
MedComm (2020), 5, 2024
|
|
8HLD
 
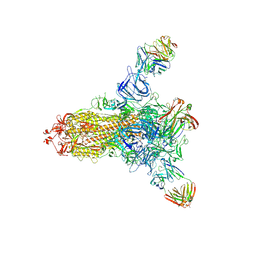 | | S protein of SARS-CoV-2 in complex with 26434 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhang, Y.Y, Guo, Y.Y, Zhou, Q. | | Deposit date: | 2022-11-29 | | Release date: | 2024-06-05 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Defining the features and structure of neutralizing antibody targeting the silent face of the SARS-CoV-2 spike N-terminal domain.
MedComm (2020), 5, 2024
|
|
6LK5
 
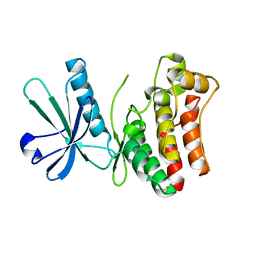 | | MLKL mutant - T357ES358D | | Descriptor: | Mixed lineage kinase domain-like protein | | Authors: | Wang, H.Y, Li, S, Zhang, Y. | | Deposit date: | 2019-12-18 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The MLKL kinase-like domain dimerization is an indispensable step of mammalian MLKL activation in necroptosis signaling.
Cell Death Dis, 12, 2021
|
|
6LK6
 
 | | MLKL mutant - T357AS358A | | Descriptor: | Mixed lineage kinase domain-like protein | | Authors: | Wang, H, Li, S, Zhang, Y. | | Deposit date: | 2019-12-18 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The MLKL kinase-like domain dimerization is an indispensable step of mammalian MLKL activation in necroptosis signaling.
Cell Death Dis, 12, 2021
|
|
6DQB
 
 | | LINKED KDM5A JMJ DOMAIN FORMING COVALENT BOND TO INHIBITOR N71 i.e. 2-((3-(4-(dimethylamino)but-2-enamido)phenyl)(2-(piperidin-1-yl)ethoxy)methyl)thieno[3,2-b]pyridine-7-carboxylic acid | | Descriptor: | 2-{(R)-(3-{[(2E)-4-(dimethylamino)but-2-enoyl]amino}phenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, 2-{(R)-(3-{[4-(dimethylamino)butanoyl]amino}phenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, 2-{(S)-(3-{[4-(dimethylamino)butanoyl]amino}phenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
6DQ6
 
 | |
6DQF
 
 | |
6DQA
 
 | | Linked KDM5A JMJ Domain Bound to Inhibitor N70 i.e.[2-((3-aminophenyl)(2-(piperidin-1-yl)ethoxy)methyl)thieno[3,2-b]pyridine-7-carboxylic acid] | | Descriptor: | 1,2-ETHANEDIOL, 2-{(R)-(3-aminophenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.888 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
6DQ9
 
 | | Linked KDM5A JMJ Domain Bound to the Covalent Inhibitor N69 i.e. [2-((3-acrylamidophenyl)(2-(piperidin-1-yl)ethoxy)methyl)thieno[3,2-b]pyridine-7-carboxylic acid] | | Descriptor: | 1,2-ETHANEDIOL, 2-{(R)-[3-(acryloylamino)phenyl][2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
6DQ5
 
 | |
6DQE
 
 | |
6DQD
 
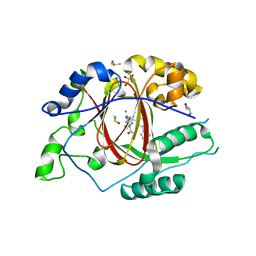 | |
6DQ4
 
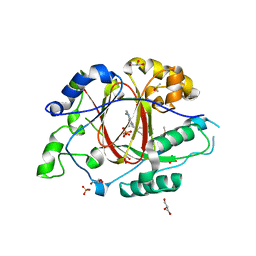 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR GSK-J1 | | Descriptor: | 3-[[2-pyridin-2-yl-6-(1,2,4,5-tetrahydro-3-benzazepin-3-yl)pyrimidin-4-yl]amino]propanoic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.392 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
6DQ8
 
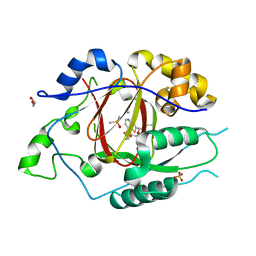 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR N49 i.e. 2-((2-chlorophenyl)(2-(1-methylpyrrolidin-2-yl)ethoxy)methyl)thieno[3,2-b]pyridine-7-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-[(R)-(2-chlorophenyl){2-[(2S)-1-methylpyrrolidin-2-yl]ethoxy}methyl]thieno[3,2-b]pyridine-7-carboxylic acid, 2-[(S)-(2-chlorophenyl){2-[(2S)-1-methylpyrrolidin-2-yl]ethoxy}methyl]thieno[3,2-b]pyridine-7-carboxylic acid, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
6DQ7
 
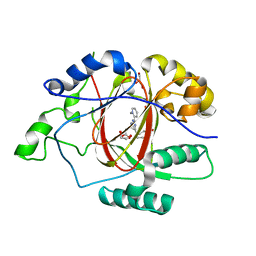 | |
6DQC
 
 | |
6RJK
 
 | | Structure of virulence factor SghA from Agrobacterium tumefaciens | | Descriptor: | Beta-glucosidase | | Authors: | Ye, F.Z, Wang, C, Chang, C.Q, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2019-04-27 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Agrobacteria reprogram virulence gene expression by controlled release of host-conjugated signals.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6RK2
 
 | | Complex structure of virulence factor SghA mutant with its substrate SAG | | Descriptor: | 2-(alpha-L-altropyranosyloxy)benzoic acid, Beta-glucosidase | | Authors: | Ye, F.Z, Wang, C, Chang, C.Q, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2019-04-30 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Agrobacteria reprogram virulence gene expression by controlled release of host-conjugated signals.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6RJM
 
 | | Complex structure of virulence factor SghA and its hydrolysis product glucose | | Descriptor: | Beta-glucosidase, alpha-D-glucopyranose | | Authors: | Ye, F.Z, Wang, C, Chang, C.Q, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2019-04-27 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | Agrobacteria reprogram virulence gene expression by controlled release of host-conjugated signals.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6RJO
 
 | | Complex structure of virulence factor SghA with its substrate analog salicin | | Descriptor: | 2-(hydroxymethyl)phenyl beta-D-glucopyranoside, Beta-glucosidase | | Authors: | Ye, F.Z, Wang, C, Chang, C.Q, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2019-04-28 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Agrobacteria reprogram virulence gene expression by controlled release of host-conjugated signals.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6S7V
 
 | | Lipoteichoic acids flippase LtaA | | Descriptor: | MFS transporter | | Authors: | Zhang, B, Perez, C. | | Deposit date: | 2019-07-06 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | Structure of a proton-dependent lipid transporter involved in lipoteichoic acids biosynthesis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
