8DUN
 
 | | Cryo-EM Structure of Antibody SKW11 in complex with Western Equine Encephalitis Virus spike (local refinement from VLP particles) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody SKW11 heavy chain, ... | | Authors: | Cerutti, G, Verardi, R, Roederer, M, Shapiro, L. | | Deposit date: | 2022-07-27 | | Release date: | 2023-07-05 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (5.84 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
8DEE
 
 | | Asymmetric Unit of Western Equine Encephalitis Virus | | Descriptor: | Capsid protein, Spike glycoprotein E1, Spike glycoprotein E2 | | Authors: | Pletnev, S, Verardi, R, Roedeger, M, Kwong, P. | | Deposit date: | 2022-06-20 | | Release date: | 2023-07-05 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
8DEF
 
 | | Cryo-EM Structure of Western Equine Encephalitis Virus VLP in complex with SKW24 fab | | Descriptor: | SKW24 Fab heavy chain, SKW24 Fab light chain, Spike glycoprotein E1, ... | | Authors: | Pletnev, S, Tsybovsky, Y, Verardi, R, Roedeger, M, Kwong, P.D. | | Deposit date: | 2022-06-20 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
8DWO
 
 | | Cryo-EM Structure of Eastern Equine Encephalitis Virus in complex with SKE26 Fab | | Descriptor: | Envelope glycoprotein E1, Envelope glycoprotein E2, SKE26 Fab Heavy Chain, ... | | Authors: | Pletnev, S, Verardi, R, Roedeger, M, Kwong, P. | | Deposit date: | 2022-08-01 | | Release date: | 2023-07-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
5ESZ
 
 | | Crystal Structure of Broadly Neutralizing Antibody CH04, Isolated from Donor CH0219, in Complex with Scaffolded Trimeric HIV-1 Env V1V2 Domain from the Clade AE Strain A244 | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase,Envelope glycoprotein gp160, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH04 Heavy Chain, ... | | Authors: | Gorman, J, Yang, M, Kwong, P.D. | | Deposit date: | 2015-11-17 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4.191 Å) | | Cite: | Structures of HIV-1 Env V1V2 with broadly neutralizing antibodies reveal commonalities that enable vaccine design.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5ESV
 
 | | Crystal Structure of Broadly Neutralizing Antibody CH03, Isolated from Donor CH0219, in Complex with Scaffolded Trimeric HIV-1 Env V1V2 Domain from the Clade C Superinfecting Strain of Donor CAP256. | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase,Envelope glycoprotein gp160, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorman, J, Yang, M, Kwong, P.D. | | Deposit date: | 2015-11-17 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.105 Å) | | Cite: | Structures of HIV-1 Env V1V2 with broadly neutralizing antibodies reveal commonalities that enable vaccine design.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5FYL
 
 | | Crystal Structure at 3.7 A Resolution of Fully Glycosylated HIV-1 Clade A BG505 SOSIP.664 Prefusion Env Trimer in Complex with Broadly Neutralizing Antibodies PGT122 and 35O22 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 ANTIBODY FAB HEAVY CHAIN, ... | | Authors: | Stewart-Jones, G.B.E, Zhou, T, Thomas, P.V, Kwong, P.D. | | Deposit date: | 2016-03-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B and G
Cell(Cambridge,Mass.), 165, 2016
|
|
5FYJ
 
 | | Crystal Structure at 3.4 A Resolution of Fully Glycosylated HIV-1 Clade G X1193.c1 SOSIP.664 Prefusion Env Trimer in Complex with Broadly Neutralizing Antibodies PGT122, 35O22 and VRC01 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stewart-Jones, G.B.E, Zhou, T, Thomas, P.V, Kwong, P.D. | | Deposit date: | 2016-03-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B and G
Cell(Cambridge,Mass.), 165, 2016
|
|
5F96
 
 | |
8EEU
 
 | | Venezuelan equine encephalitis virus-like particle in complex with Fab SKT05 | | Descriptor: | Coat protein, Fab SKT05 heavy chain, Fab SKT05 light chain | | Authors: | Tsybovsky, Y, Pletnev, S, Verardi, R, Roederer, M, Kwong, P.D. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-19 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
8EEV
 
 | | Venezuelan equine encephalitis virus-like particle in complex with Fab SKT-20 | | Descriptor: | Coat protein, Fab SKT20 heavy chain, Fab SKT20 light chain | | Authors: | Tsybovsky, Y, Pletnev, S, Verardi, R, Roederer, M, Kwong, P.D. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
5F9W
 
 | | Crystal structure of broadly neutralizing VH1-46 germline-derived CD4-binding site-directed antibody CH235 in complex with HIV-1 clade A/E 93TH057 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of CH235-lineage antibody CH235, Light chain of CH235-lineage antibody CH235, ... | | Authors: | Chen, L, Zhou, T, Kwong, P.D. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-09 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.8911 Å) | | Cite: | Maturation Pathway from Germline to Broad HIV-1 Neutralizer of a CD4-Mimic Antibody.
Cell, 165, 2016
|
|
5FYK
 
 | | Crystal Structure at 3.7 A Resolution of Fully Glycosylated HIV-1 Clade B JR-FL SOSIP.664 Prefusion Env Trimer in Complex with Broadly Neutralizing Antibodies PGT122, 35O22 and VRC01 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22, ... | | Authors: | Stewart-Jones, G.B.E, Zhou, T, Thomas, P.V, Kwong, P.D. | | Deposit date: | 2016-03-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.107 Å) | | Cite: | Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B and G
Cell(Cambridge,Mass.), 165, 2016
|
|
8FLW
 
 | | Cryo-EM Structure of PGT145 DU303 Fab in complex with BG505 DS-SOSIP.664 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2022-12-22 | | Release date: | 2023-05-31 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Improved HIV-1 neutralization breadth and potency of V2-apex antibodies by in silico design.
Cell Rep, 42, 2023
|
|
8FK5
 
 | | Cryo-EM Structure of PG9RSH DU011 Fab in complex with BG505 DS-SOSIP.664 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2022-12-20 | | Release date: | 2023-05-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Improved HIV-1 neutralization breadth and potency of V2-apex antibodies by in silico design.
Cell Rep, 42, 2023
|
|
8FL1
 
 | | Cryo-EM Structure of PG9RSH DU025 Fab in complex with BG505 DS-SOSIP.664 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2022-12-21 | | Release date: | 2023-05-31 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Improved HIV-1 neutralization breadth and potency of V2-apex antibodies by in silico design.
Cell Rep, 42, 2023
|
|
8XGS
 
 | | a peptide receptor complex structure | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Wu, Z, Du, Y, Chen, G. | | Deposit date: | 2023-12-15 | | Release date: | 2024-10-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis for the ligand recognition and G protein subtype selectivity of kisspeptin receptor.
Sci Adv, 10, 2024
|
|
8XGO
 
 | | a peptide receptor complex structure | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Wu, Z, Du, Y, Chen, G. | | Deposit date: | 2023-12-15 | | Release date: | 2024-10-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural basis for the ligand recognition and G protein subtype selectivity of kisspeptin receptor.
Sci Adv, 10, 2024
|
|
8FEN
 
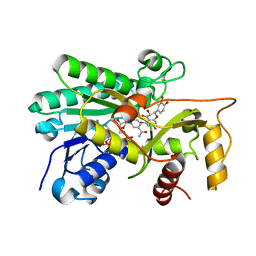 | | Panicum vigratum Dihydroflavonol 4-reductase complexed with NADP and DHQ | | Descriptor: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Panicum virgatum Dihydroflavonol 4-Reductase | | Authors: | Lewis, J.A, Kang, C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
8FEM
 
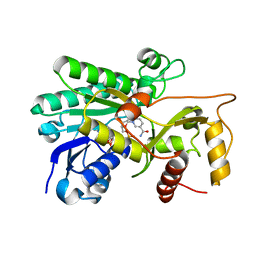 | | Panicum vigratum Dihydroflavonol 4-reductase complexed with NADP | | Descriptor: | Dihydroflavonol 4-Reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lewis, J.A, Kang, C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
8F7T
 
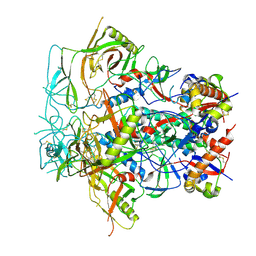 | | Glycan-Base ConC Env Trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 Env gp120, ... | | Authors: | Olia, A.S, Kwong, P.D. | | Deposit date: | 2022-11-20 | | Release date: | 2023-09-27 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Soluble prefusion-closed HIV-envelope trimers with glycan-covered bases.
Iscience, 26, 2023
|
|
8XGU
 
 | | a peptide receptor complex structure | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wu, Z, Du, Y, Chen, G. | | Deposit date: | 2023-12-15 | | Release date: | 2024-10-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the ligand recognition and G protein subtype selectivity of kisspeptin receptor.
Sci Adv, 10, 2024
|
|
8ZCF
 
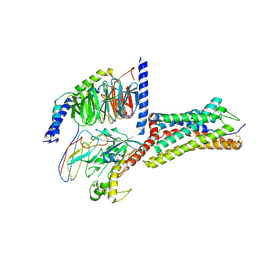 | | Cryo-EM structure of GPR4 complexed with Gs in pH7.5 | | Descriptor: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yue, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-04-29 | | Release date: | 2025-04-23 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of stepwise proton sensing-mediated GPCR activation.
Cell Res., 35, 2025
|
|
8ZCE
 
 | | Cryo-EM structure of GPR4 complexed with Gs in pH6.0 | | Descriptor: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yue, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-04-29 | | Release date: | 2025-04-23 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of stepwise proton sensing-mediated GPCR activation.
Cell Res., 35, 2025
|
|
5F9O
 
 | | Crystal structure of broadly neutralizing VH1-46 germline-derived CD4-binding site-directed antibody CH235.09 in complex with HIV-1 clade A/E 93TH057 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CH235.09 Light chain, ... | | Authors: | Joyce, M.G, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Maturation Pathway from Germline to Broad HIV-1 Neutralizer of a CD4-Mimic Antibody.
Cell, 165, 2016
|
|
