8WU3
 
 | |
8WU2
 
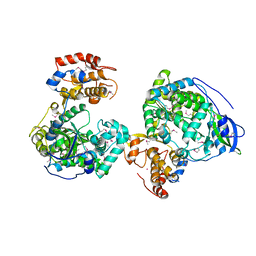 | | Crystal structure of RNA-dependent RNA polymerases from Jingmen tick virus | | Descriptor: | RNA polymerase, SELENIUM ATOM | | Authors: | Liu, Z, Han, P, Peng, Q, Qi, J. | | Deposit date: | 2023-10-20 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of RNA-dependent RNA polymerases from Jingmen tick virus and Alongshan virus
HLIFE, 2, 2024
|
|
6M6C
 
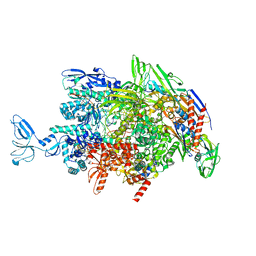 | | CryoEM structure of Thermus thermophilus RNA polymerase elongation complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2020-03-14 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
7QWM
 
 | |
7QVF
 
 | |
7QVC
 
 | |
7QWL
 
 | |
7QWG
 
 | |
6M6A
 
 | | Cryo-EM structure of Thermus thermophilus Mfd in complex with RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2020-03-14 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
8HJU
 
 | | Cryo-EM structure of native RC-LH complex from Roseiflexus castenholzii at 10,000 lux | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-methyl-3-[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,1 4,18,22,26,30,34,38,42-undecaen-1-yl]naphthalene-1,4-dione, Alpha subunit of light-harvesting 1, ... | | Authors: | Xu, X, Xin, J. | | Deposit date: | 2022-11-23 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Carotenoid assembly regulates quinone diffusion and the Roseiflexus castenholzii reaction center-light harvesting complex architecture.
Elife, 12, 2023
|
|
8HJV
 
 | | Cryo-EM structure of carotenoid-depleted RC-LH complex from Roseiflexus castenholzii at 10,000 lux | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-methyl-3-[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,1 4,18,22,26,30,34,38,42-undecaen-1-yl]naphthalene-1,4-dione, Alpha subunit of light-harvesting 1, ... | | Authors: | Xu, X, Xin, J. | | Deposit date: | 2022-11-23 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Carotenoid assembly regulates quinone diffusion and the Roseiflexus castenholzii reaction center-light harvesting complex architecture.
Elife, 12, 2023
|
|
8IPI
 
 | | The apo structure of human mitochondrial methyltransferase METTL15 | | Descriptor: | 12S rRNA N4-methylcytidine (m4C) methyltransferase | | Authors: | Lv, M.Q, Zhou, W.W. | | Deposit date: | 2023-03-14 | | Release date: | 2024-01-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the specific recognition of mitochondrial ribosome-binding factor hsRBFA and 12 S rRNA by methyltransferase METTL15.
Cell Discov, 10, 2024
|
|
2Y1R
 
 | | Structure of MecA121 & ClpC N-domain complex | | Descriptor: | ADAPTER PROTEIN MECA 1, NEGATIVE REGULATOR OF GENETIC COMPETENCE CLPC/MECB, S,R MESO-TARTARIC ACID | | Authors: | Wang, F, Mei, Z.Q, Wang, J.W, Shi, Y.G. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Structure and Mechanism of the Hexameric Meca-Clpc Molecular Machine.
Nature, 471, 2011
|
|
2Y1Q
 
 | | Crystal Structure of ClpC N-terminal Domain | | Descriptor: | NEGATIVE REGULATOR OF GENETIC COMPETENCE CLPC/MECB, SULFATE ION | | Authors: | Wang, F, Mei, Z.Q, Wang, J.W, Shi, Y.G. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Mechanism of the Hexameric Meca-Clpc Molecular Machine.
Nature, 471, 2011
|
|
7BSD
 
 | |
7BSC
 
 | | Complex structure of 1G5.3 Fab bound to DENV2 NS1c | | Descriptor: | 1G5.3 Fab Heavy Chain, 1G5.3 Fab Light Chain, Non-structural protein 1 | | Authors: | Song, H, Qi, J, Gao, F.G. | | Deposit date: | 2020-03-30 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A broadly protective antibody that targets the flavivirus NS1 protein.
Science, 371, 2021
|
|
7ZC4
 
 | | Cryo-EM structure of POLRMT mutant. | | Descriptor: | DNA-directed RNA polymerase, mitochondrial | | Authors: | Das, H, Hallberg, B.M. | | Deposit date: | 2022-03-25 | | Release date: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Non-coding 7S RNA inhibits transcription via mitochondrial RNA polymerase dimerization.
Cell, 185, 2022
|
|
4DDP
 
 | |
6L1P
 
 | | Crystal structure of PHF20L1 in complex with Hit 1 | | Descriptor: | 4-(1-methyl-3,6-dihydro-2H-pyridin-4-yl)phenol, GLYCEROL, PHD finger protein 20-like protein 1, ... | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-29 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.231 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1C
 
 | | Crystal Structure Of of PHF20L1 Tudor1 Y24L mutant | | Descriptor: | GLYCEROL, PHD finger protein 20-like protein 1, SULFATE ION | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-28 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1I
 
 | | Crystal Structure Of of PHF20L1 Tudor1 Y24W/Y29W mutant | | Descriptor: | PHD finger protein 20-like protein 1, SULFATE ION | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-29 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L10
 
 | | PHF20L1 Tudor1 - MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PHD finger protein 20-like protein 1, SULFATE ION | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-27 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1F
 
 | |
7RTC
 
 | |
5XFN
 
 | | Structure of the N-terminal domains of PHF1 | | Descriptor: | PHD finger protein 1, ZINC ION | | Authors: | Wang, Z, Li, H. | | Deposit date: | 2017-04-11 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Polycomb-like proteins link the PRC2 complex to CpG islands
Nature, 549, 2017
|
|
