4RDJ
 
 | | Crystal structure of Norovirus Boxer P domain | | Descriptor: | Capsid | | Authors: | Hao, N, Chen, Y, Xia, M, Liu, W, Tan, M, Jiang, X, Li, X. | | Deposit date: | 2014-09-19 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of GI.8 Boxer virus P dimers in complex with HBGAs, a novel evolutionary path selected by the Lewis epitope.
Protein Cell, 6, 2015
|
|
4S1T
 
 | |
8IF8
 
 | | Arabinosyltransferase AftA | | Descriptor: | CALCIUM ION, Galactan 5-O-arabinofuranosyltransferase | | Authors: | Gong, Y.C, Rao, Z.H, Zhang, L. | | Deposit date: | 2023-02-17 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the priming arabinosyltransferase AftA required for AG biosynthesis of Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6KGT
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with faropenem | | Descriptor: | (2R)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-5-[(2R)-oxolan-2-yl]-2,3-dihydro-1,3-thiazole-4-carboxylic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.308 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
2Q12
 
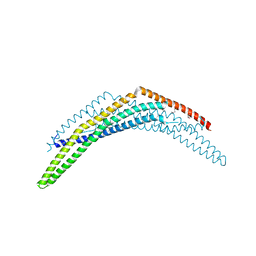 | | Crystal Structure of BAR domain of APPL1 | | Descriptor: | DCC-interacting protein 13 alpha | | Authors: | Zhang, X.C, Zhu, G. | | Deposit date: | 2007-05-23 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure of the APPL1 BAR-PH domain and characterization of its interaction with Rab5.
Embo J., 26, 2007
|
|
2Q13
 
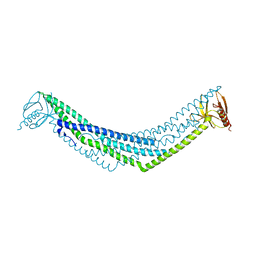 | | Crystal structure of BAR-PH domain of APPL1 | | Descriptor: | DCC-interacting protein 13 alpha | | Authors: | Zhu, G, Zhang, X.C. | | Deposit date: | 2007-05-23 | | Release date: | 2007-08-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the APPL1 BAR-PH domain and characterization of its interaction with Rab5.
Embo J., 26, 2007
|
|
8HCZ
 
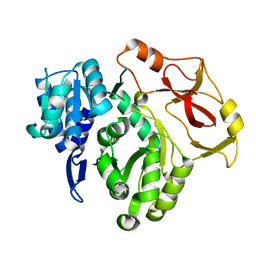 | |
8HDF
 
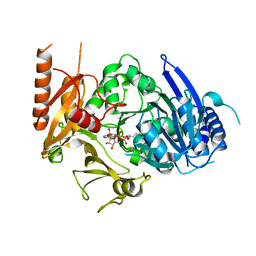 | | Full length crystal structure of mycobacterium tuberculosis FadD23 in complex with ANP and PLM | | Descriptor: | Long-chain-fatty-acid--AMP ligase FadD23, PALMITIC ACID, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yan, M.R, Liu, X, Zhang, W, Rao, Z.H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The Key Roles of Mycobacterium tuberculosis FadD23 C-terminal Domain in Catalytic Mechanisms.
Front Microbiol, 14, 2023
|
|
8HD4
 
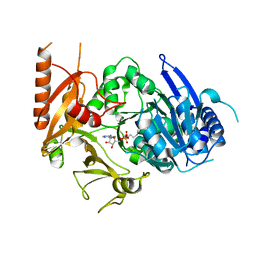 | | Full-length crystal structure of mycobacterium tuberculosis FadD23 in complex with AMPC16 | | Descriptor: | Long-chain-fatty-acid--AMP ligase FadD23, palmitoyl adenylate | | Authors: | Yan, M.R, Liu, X, Zhang, W, Rao, Z.H. | | Deposit date: | 2022-11-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | The Key Roles of Mycobacterium tuberculosis FadD23 C-terminal Domain in Catalytic Mechanisms.
Front Microbiol, 14, 2023
|
|
5ZUD
 
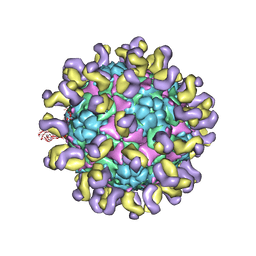 | | Fit R10 Fab coordinates into the cryo-EM of EV71 in complex with D6 | | Descriptor: | Capsid protein VP1, R10 ANTIBODY HEAVY CHAIN, R10 ANTIBODY LIGHT CHAIN, ... | | Authors: | Wang, X, Zhu, L, Wang, N. | | Deposit date: | 2018-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Neutralization Mechanisms of Two Highly Potent Antibodies against Human Enterovirus 71.
Mbio, 9, 2018
|
|
5ZV7
 
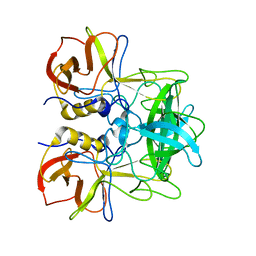 | | P domain of GII.17-2014/15 complexed with B-trisaccharide | | Descriptor: | VP1, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]alpha-D-galactopyranose | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-09 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
5ZVC
 
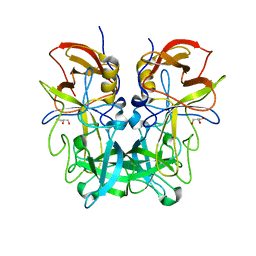 | | P domain of GII.13 norovirus capsid complexed with Lewis A trisaccharide | | Descriptor: | GLYCEROL, Major capsid protein VP1, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-10 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
5ZV9
 
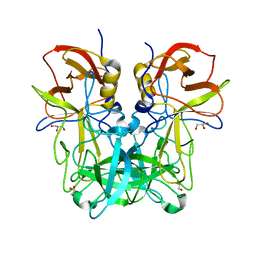 | | P domain of GII.13 norovirus capsid | | Descriptor: | GLYCEROL, Major capsid protein VP1 | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-09 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
5ZUS
 
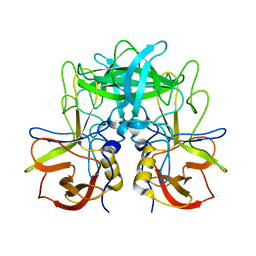 | | P domain of GII.17-2014/15 | | Descriptor: | VP1 | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-08 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
5ZUF
 
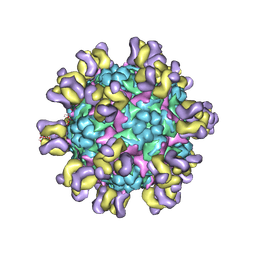 | | Fit R10 Fab coordinates into the cryo-EM of EV71 in complex with A9 | | Descriptor: | Capsid protein VP1, R10 ANTIBODY HEAVY CHAIN, R10 ANTIBODY LIGHT CHAIN, ... | | Authors: | Wang, X, Zhu, L, Wang, N. | | Deposit date: | 2018-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Neutralization Mechanisms of Two Highly Potent Antibodies against Human Enterovirus 71.
Mbio, 9, 2018
|
|
5ZV5
 
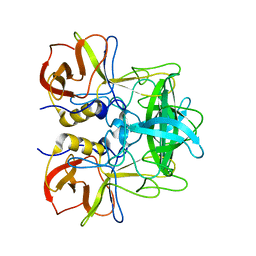 | | P domain of GII.17-2014/15 complexed with A-trisaccharide | | Descriptor: | VP1, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]alpha-D-galactopyranose | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-09 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
5ZKQ
 
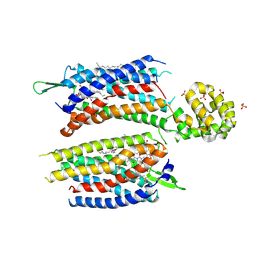 | | Crystal structure of the human platelet-activating factor receptor in complex with ABT-491 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-ethynyl-3-{3-fluoro-4-[(2-methyl-1H-imidazo[4,5-c]pyridin-1-yl)methyl]benzene-1-carbonyl}-N,N-dimethyl-1H-indole-1-carboxamide, Platelet-activating factor receptor,Endolysin,Endolysin,Platelet-activating factor receptor, ... | | Authors: | Cao, C, Zhao, Q, Zhang, X.C, Wu, B. | | Deposit date: | 2018-03-25 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for signal recognition and transduction by platelet-activating-factor receptor.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5ZKP
 
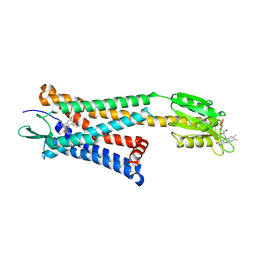 | | Crystal structure of the human platelet-activating factor receptor in complex with SR 27417 | | Descriptor: | FLAVIN MONONUCLEOTIDE, N1,N1-dimethyl-N2-[(pyridin-3-yl)methyl]-N2-{4-[2,4,6-tri(propan-2-yl)phenyl]-1,3-thiazol-2-yl}ethane-1,2-diamine, Platelet-activating factor receptor,Flavodoxin,Platelet-activating factor receptor | | Authors: | Cao, C, Zhao, Q, Zhang, X.C, Wu, B. | | Deposit date: | 2018-03-25 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis for signal recognition and transduction by platelet-activating-factor receptor.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5ZUQ
 
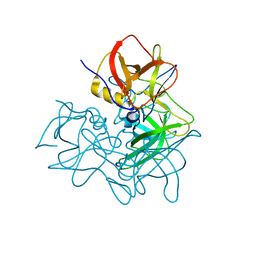 | | P domain of GII.17-1978 | | Descriptor: | VP1 | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-08 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
2FBW
 
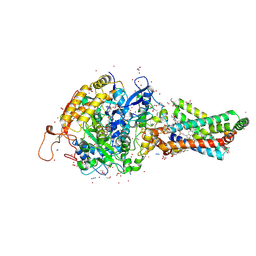 | | Avian respiratory complex II with carboxin bound | | Descriptor: | (~{Z})-2-oxidanylbut-2-enedioic acid, 2-METHYL-N-PHENYL-5,6-DIHYDRO-1,4-OXATHIINE-3-CARBOXAMIDE, AZIDE ION, ... | | Authors: | Huang, L.S, Sun, G, Cobessi, D, Wang, A.C, Shen, J.T, Tung, E.Y, Anderson, V.E, Berry, E.A. | | Deposit date: | 2005-12-10 | | Release date: | 2005-12-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | 3-nitropropionic acid is a suicide inhibitor of mitochondrial respiration that, upon oxidation by complex II, forms a covalent adduct with a catalytic base arginine in the active site of the enzyme.
J.Biol.Chem., 281, 2006
|
|
8WON
 
 | | Cryo-EM structure of the 10-subunits Mmp1 complex from Mycobacterium smegmatis | | Descriptor: | Major membrane protein I | | Authors: | Zhang, M, Tang, Y, Gao, Y, Liu, X, Lan, W, Liu, Y, Ma, M. | | Deposit date: | 2023-10-07 | | Release date: | 2025-01-01 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | The structural and functional analysis of mycobacteria cysteine desulfurase-loaded encapsulin.
Commun Biol, 7, 2024
|
|
7CY2
 
 | |
7CZ2
 
 | |
7CYR
 
 | |
7WS1
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 510A5 heavy chain, ... | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
