7XX1
 
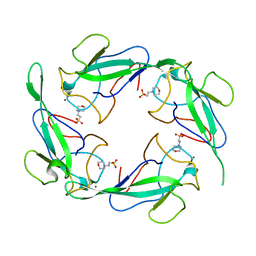 | | Crystal structure of SARS-CoV-2 N-NTD | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nucleoprotein, ZINC ION | | Authors: | Luan, X.D, Li, X.M, Li, Y.F. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antiviral drug design based on structural insights into the N-terminal domain and C-terminal domain of the SARS-CoV-2 nucleocapsid protein.
Sci Bull (Beijing), 67, 2022
|
|
4Z03
 
 | | C. bescii Family 3 pectate lyase double mutant K108A in complex with trigalacturonic acid | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2015-03-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The catalytic mechanism and unique low pH optimum of Caldicellulosiruptor bescii family 3 pectate lyase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4YZX
 
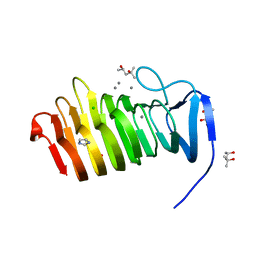 | | C. bescii Family 3 pectate lyase double mutant K108A/D107N in complex with trigalacturonic acid | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2015-03-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The catalytic mechanism and unique low pH optimum of Caldicellulosiruptor bescii family 3 pectate lyase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7XWZ
 
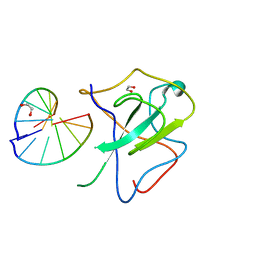 | | Crystal structure of SARS-CoV-2 N-NTD and dsRNA complex | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Nucleoprotein, ... | | Authors: | Luan, X.D, Li, X.M, Li, Y.F. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Antiviral drug design based on structural insights into the N-terminal domain and C-terminal domain of the SARS-CoV-2 nucleocapsid protein.
Sci Bull (Beijing), 67, 2022
|
|
7XWX
 
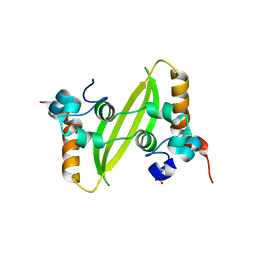 | | Crystal structure of SARS-CoV-2 N-CTD | | Descriptor: | Nucleoprotein, PHOSPHATE ION | | Authors: | Luan, X.D, Li, X.M, Li, Y.F. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Antiviral drug design based on structural insights into the N-terminal domain and C-terminal domain of the SARS-CoV-2 nucleocapsid protein.
Sci Bull (Beijing), 67, 2022
|
|
7BNR
 
 | |
7BNK
 
 | |
3DX5
 
 | | Crystal structure of the probable 3-DHS dehydratase AsbF involved in the petrobactin synthesis from Bacillus anthracis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,4-DIHYDROXYBENZOIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Maltseva, N, Stols, L, Eschenfeldt, W, Pfleger, B.F, Sherman, D.H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-23 | | Release date: | 2008-09-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural and functional analysis of AsbF: origin of the stealth 3,4-dihydroxybenzoic acid subunit for petrobactin biosynthesis.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
8AYT
 
 | | Crystal structure of SUDV VP40 W95A mutant | | Descriptor: | Matrix protein VP40 | | Authors: | Werner, A.-D, Steinchen, W, Werel, L, Kowalski, K, Essen, L.-O, Becker, S. | | Deposit date: | 2022-09-03 | | Release date: | 2023-09-13 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of SUDV VP40 W95A mutant
To Be Published
|
|
8AYU
 
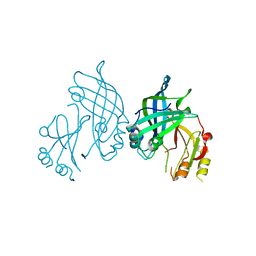 | | Crystal structure of SUDV VP40 L117A mutant | | Descriptor: | Matrix protein VP40 | | Authors: | Werner, A.-D, Steinchen, W, Werel, L, Kowalski, K, Essen, L.-O, Becker, S. | | Deposit date: | 2022-09-03 | | Release date: | 2023-09-13 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of SUDV VP40 L117A mutant
To Be Published
|
|
6J46
 
 | | LepI-SAH complex structure | | Descriptor: | O-methyltransferase lepI, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Qiu, S, Wei, C. | | Deposit date: | 2019-01-08 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.621 Å) | | Cite: | Deciphering the regulatory and catalytic mechanisms of an unusual SAM-dependent enzyme.
Signal Transduct Target Ther, 4, 2019
|
|
6J9E
 
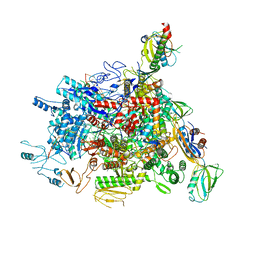 | |
6J24
 
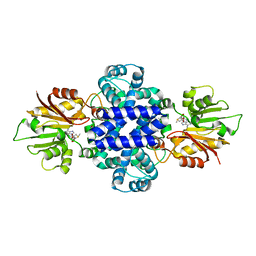 | | Crystal structure of a SAM-dependent methyltransferase LepI in complex with its substrate | | Descriptor: | (3~{S},4'~{R},4'~{a}~{S},6'~{R},8'~{a}~{S})-4',6'-dimethyl-5-phenyl-spiro[1~{H}-pyridine-3,5'-2,3,4,4~{a},6,8~{a}-hexahydro-1~{H}-naphthalene]-2,4-dione, O-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Qiu, S, Wei, C. | | Deposit date: | 2018-12-30 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Deciphering the regulatory and catalytic mechanisms of an unusual SAM-dependent enzyme.
Signal Transduct Target Ther, 4, 2019
|
|
6J9F
 
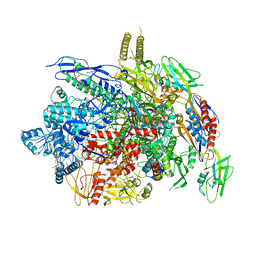 | |
6J1O
 
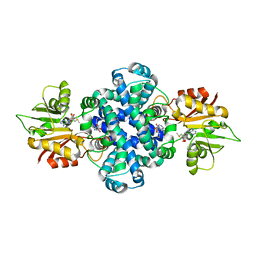 | |
6CP9
 
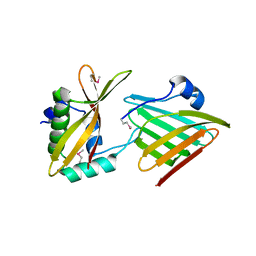 | | Contact-dependent growth inhibition toxin - immunity protein complex from Klebsiella pneumoniae 342 | | Descriptor: | CdiA, CdiI | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Convergent Evolution of the Barnase/EndoU/Colicin/RelE (BECR) Fold in Antibacterial tRNase Toxins.
Structure, 27, 2019
|
|
5HKQ
 
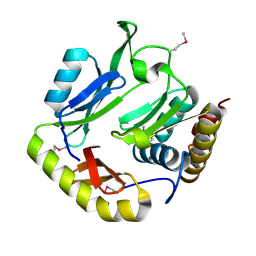 | | Crystal structure of CDI complex from Escherichia coli STEC_O31 | | Descriptor: | CdiI immunity protein, Contact-dependent inhibitor A | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2020-03-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional plasticity of antibacterial EndoU toxins.
Mol.Microbiol., 109, 2018
|
|
6CP8
 
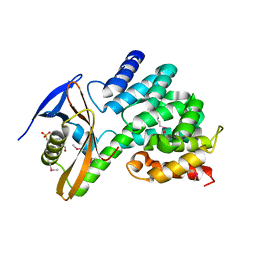 | | Contact-dependent growth inhibition toxin-immunity protein complex from from E. coli 3006 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CdiA, CdiI, ... | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Convergent Evolution of the Barnase/EndoU/Colicin/RelE (BECR) Fold in Antibacterial tRNase Toxins.
Structure, 27, 2019
|
|
5I4R
 
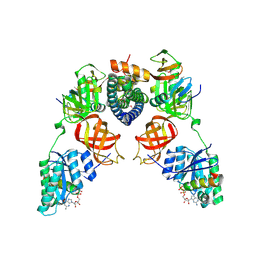 | | Contact-dependent inhibition system from Escherichia coli NC101 - ternary CdiA/CdiI/EF-Tu complex (trypsin-modified) | | Descriptor: | Contact-dependent inhibitor A, Contact-dependent inhibitor I, Elongation factor Tu, ... | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2016-02-12 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of a novel antibacterial toxin that exploits elongation factor Tu to cleave specific transfer RNAs.
Nucleic Acids Res., 45, 2017
|
|
5I4Q
 
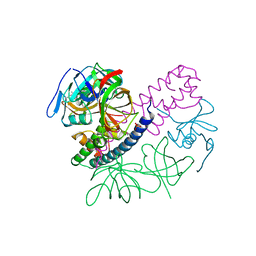 | | Contact-dependent inhibition system from Escherichia coli NC101 - ternary CdiA/CdiI/EF-Tu complex (domains 2 and 3) | | Descriptor: | CHLORIDE ION, Contact-dependent inhibitor A, Contact-dependent inhibitor I, ... | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2016-02-12 | | Release date: | 2017-06-28 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of a novel antibacterial toxin that exploits elongation factor Tu to cleave specific transfer RNAs.
Nucleic Acids Res., 45, 2017
|
|
3LFM
 
 | |
7XR1
 
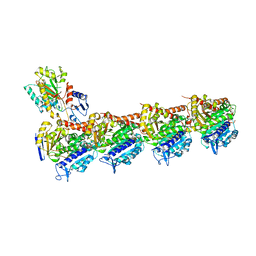 | | Crystal structure of T2R-TTL-3a complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloranyl-6-fluoranyl-N-(4-methoxyphenyl)-N-methyl-quinazolin-4-amine, CALCIUM ION, ... | | Authors: | Lun, T, Wu, C.Y. | | Deposit date: | 2022-05-09 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structure of T2R-TTL-3a complex
To Be Published
|
|
7XR0
 
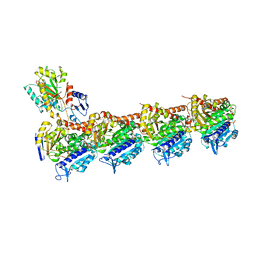 | | Crystal structure of T2R-TTL-27a complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloranyl-5-fluoranyl-N-(4-methoxyphenyl)-N-methyl-quinazolin-4-amine, CALCIUM ION, ... | | Authors: | Lun, T, Wu, C.Y. | | Deposit date: | 2022-05-09 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of T2R-TTL-27a complex
To Be Published
|
|
7AXU
 
 | |
7AXQ
 
 | |
