4XV0
 
 | | Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Trichoderma reesei | | Descriptor: | Beta-xylanase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Savchenko, A. | | Deposit date: | 2015-01-26 | | Release date: | 2015-02-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9697 Å) | | Cite: | Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Trichoderma reesei
To Be Published
|
|
2O6B
 
 | | Crystal structure of the PA5185 protein from Pseudomonas Aeruginosa strain PAO1- new crystal form. | | Descriptor: | THIOESTERASE | | Authors: | Chruszcz, M, Koclega, K.D, Evdokimova, E, Cymborowski, M, Kudritska, M, Savchenko, A, Edwards, A, Minor, W. | | Deposit date: | 2006-12-07 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Function-biased choice of additives for optimization of protein crystallization - the case of the putative thioesterase PA5185 from Pseudomonas aeruginosa PAO1.
Cryst.Growth Des., 8, 2008
|
|
2O6U
 
 | | Crystal structure of the PA5185 protein from Pseudomonas Aeruginosa strain PAO1- new crystal form. | | Descriptor: | THIOESTERASE | | Authors: | Chruszcz, M, Koclega, K.D, Evdokimova, E, Cymborowski, M, Kudritska, M, Savchenko, A, Edwards, A, Minor, W. | | Deposit date: | 2006-12-08 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Function-biased choice of additives for optimization of protein crystallization - the case of the putative thioesterase PA5185 from Pseudomonas aeruginosa PAO1.
Cryst.Growth Des., 8, 2008
|
|
4XX6
 
 | | Crystal structure of a glycosylated endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Gloeophyllum trabeum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-xylanase, ... | | Authors: | Stogios, P.J, Nocek, B, Xu, X, Cui, H, Lowden, M, Savchenko, A. | | Deposit date: | 2015-01-29 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a glycosylated endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Gloeophyllum trabeum.
To Be Published
|
|
3ICF
 
 | | Structure of Protein serine/threonine phosphatase from Saccharomyces cerevisiae with similarity to human phosphatase PP5 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FE (III) ION, ... | | Authors: | Singer, A.U, Xu, X, Chang, C, Cui, H, Kagan, O, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-17 | | Release date: | 2009-08-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Protein serine/threonine phosphatase from Saccharomyces cerevisiae with similarity to human phosphatase PP5
To be Published
|
|
2ESH
 
 | | Crystal Structure of Conserved Protein of Unknown Function TM0937- a Potential Transcriptional Factor | | Descriptor: | CALCIUM ION, conserved hypothetical protein TM0937 | | Authors: | Liu, Y, Bochkareva, E, Zheng, H, Xu, X, Nocek, B, Lunin, V, Edward, A, Pai, E.F, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-10-26 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Conserved Hypothetical Protein TM0937
To be Published
|
|
2P35
 
 | | Crystal structure of trans-aconitate methyltransferase from Agrobacterium tumefaciens | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, Trans-aconitate 2-methyltransferase | | Authors: | Chang, C, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-03-08 | | Release date: | 2007-04-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of trans-aconitate methyltransferase from Agrobacterium tumefaciens
To be Published
|
|
4XUV
 
 | | Crystal structure of a glycoside hydrolase family 105 (GH105) enzyme from Thielavia terrestris | | Descriptor: | GLYCEROL, Glycoside hydrolase family 105 protein | | Authors: | Stogios, P.J, Xu, X, Cui, H, Yim, V, Savchenko, A. | | Deposit date: | 2015-01-26 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.0496 Å) | | Cite: | Crystal structure of a glycoside hydrolase family 105 (GH105) enzyme from Thielavia terrestris
To Be Published
|
|
2PFS
 
 | | Crystal structure of universal stress protein from Nitrosomonas europaea | | Descriptor: | CHLORIDE ION, Universal stress protein | | Authors: | Chruszcz, M, Evdokimova, E, Cymborowski, M, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-04-05 | | Release date: | 2007-05-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional insight into the universal stress protein family.
Evol Appl, 6, 2013
|
|
3FYN
 
 | | Crystal structure from the mobile metagenome of Cole Harbour Salt Marsh: Integron Cassette Protein HFX_CASS3 | | Descriptor: | ACETATE ION, Integron gene cassette protein HFX_CASS3, MAGNESIUM ION | | Authors: | Sureshan, V, Deshpande, C.N, Harrop, S.J, Kudritska, M, Koenig, J.E, Evdokimova, E, Osipiuk, J, Edwards, A.M, Savchenko, A, Joachimiak, A, Doolittle, W.F, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-22 | | Release date: | 2009-02-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Structure from the mobile metagenome of Cole Harbour Salt Marsh: Integron Cassette Protein HFX_CASS3
To be Published
|
|
3IB3
 
 | | Crystal Structure of SACOL2612 - CocE/NonD family hydrolase from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, CocE/NonD family hydrolase, NICKEL (II) ION, ... | | Authors: | Domagalski, M.J, Chruszcz, M, Osinski, T, Skarina, T, Onopriyenko, O, Cymborowski, M, Shumilin, I.A, Savchenko, A, Edwards, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-15 | | Release date: | 2009-08-11 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of SACOL2612 - CocE/NonD family hydrolase from Staphylococcus aureus
To be Published
|
|
3III
 
 | | 1.95 Angstrom Crystal Structure of CocE/NonD family hydrolase (SACOL2612) from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, CocE/NonD family hydrolase, NICKEL (II) ION, ... | | Authors: | Osinski, T, Chruszcz, M, Domagalski, M.J, Cymborowski, M, Shumilin, I.A, Skarina, T, Onopriyenko, O, Zimmerman, M.D, Savchenko, A, Edwards, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-01 | | Release date: | 2009-08-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of CocE/NonD family hydrolase (SACOL2612) from Staphylococcus aureus
To be Published
|
|
2O5U
 
 | | Crystal structure of the PA5185 protein from Pseudomonas Aeruginosa strain PAO1- orthorhombic form (C222). | | Descriptor: | Thioesterase | | Authors: | Chruszcz, M, Wang, S, Evdokimova, E, Koclega, K.D, Kudritska, M, Savchenko, A, Edwards, A, Minor, W. | | Deposit date: | 2006-12-06 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Function-biased choice of additives for optimization of protein crystallization - the case of the putative thioesterase PA5185 from Pseudomonas aeruginosa PAO1.
Cryst.Growth Des., 8, 2008
|
|
3GEM
 
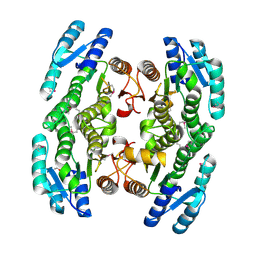 | | Crystal structure of short-chain dehydrogenase from Pseudomonas syringae | | Descriptor: | ACETATE ION, Short chain dehydrogenase | | Authors: | Osipiuk, J, Xu, X, Cui, H, Nocek, B, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-25 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | X-ray crystal structure of short-chain dehydrogenase from Pseudomonas syringae.
To be Published
|
|
4WJ0
 
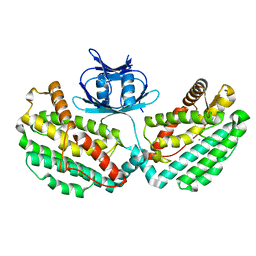 | | Structure of PH1245, a cas1 from Pyrococcus horikoshii | | Descriptor: | CHLORIDE ION, CRISPR-associated endonuclease Cas1 | | Authors: | Petit, P, Brown, G, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2014-09-29 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of PH1245, a cas1 from Pyrococcus horikoshii
To Be Published
|
|
4WQL
 
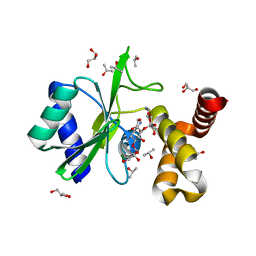 | | Crystal structure of aminoglycoside nucleotidylyltransferase ANT(2")-Ia, kanamycin-bound | | Descriptor: | 2''-aminoglycoside nucleotidyltransferase, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Cox, G, Stogios, P.J, Savchenko, A, Wright, G.D, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-22 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural and Molecular Basis for Resistance to Aminoglycoside Antibiotics by the Adenylyltransferase ANT(2)-Ia.
Mbio, 6, 2015
|
|
4HFV
 
 | | Crystal structure of lpg1851 protein from Legionella pneumophila (putative T4SS effector) | | Descriptor: | CITRIC ACID, SUCCINIC ACID, Uncharacterized protein | | Authors: | Michalska, K, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-10-05 | | Release date: | 2012-11-07 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila.
Mol Syst Biol, 12, 2016
|
|
2O8I
 
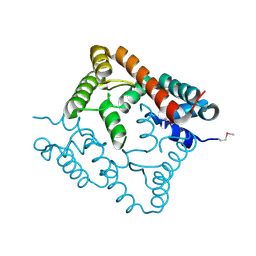 | | Crystal structure of protein Atu2327 from Agrobacterium tumefaciens str. C58 | | Descriptor: | Hypothetical protein Atu2327 | | Authors: | Chang, C, Xu, X, Gu, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-12 | | Release date: | 2007-01-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of protein Atu2327 from Agrobacterium tumefaciens str. C58
To be Published
|
|
2OB5
 
 | | Crystal structure of protein Atu2016, putative sugar binding protein | | Descriptor: | Hypothetical protein Atu2016 | | Authors: | Chang, C, Xu, X, Gu, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-18 | | Release date: | 2007-01-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of protein Atu2016, putative sugar binding protein
To be Published
|
|
2OI8
 
 | | Crystal structure of putative regulatory protein SCO4313 | | Descriptor: | Putative regulatory protein SCO4313 | | Authors: | Chang, C, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-10 | | Release date: | 2007-02-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of tetR family protein SCO4313
To be Published
|
|
2FBH
 
 | | The crystal structure of transcriptional regulator PA3341 | | Descriptor: | MERCURY (II) ION, SULFATE ION, ZINC ION, ... | | Authors: | Lunin, V.V, Evdokimova, E, Kudritska, M, Osipiuk, J, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-09 | | Release date: | 2005-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of transcriptional regulator PA3341
To be Published
|
|
3I7M
 
 | | N-terminal domain of Xaa-Pro dipeptidase from Lactobacillus brevis. | | Descriptor: | Xaa-Pro dipeptidase | | Authors: | Osipiuk, J, Xu, X, Cui, H, Ng, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-08 | | Release date: | 2009-07-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | X-ray crystal structure of N-terminal domain of Xaa-Pro dipeptidase from Lactobacillus brevis.
To be Published
|
|
4WQK
 
 | | Crystal structure of aminoglycoside nucleotidylyltransferase ANT(2")-Ia, apo form | | Descriptor: | 2''-aminoglycoside nucleotidyltransferase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Cox, G, Stogios, P.J, Savchenko, A, Wright, G.D, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-22 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Structural and Molecular Basis for Resistance to Aminoglycoside Antibiotics by the Adenylyltransferase ANT(2)-Ia.
Mbio, 6, 2015
|
|
2FFS
 
 | | Structure of PR10-allergen-like protein PA1206 from Pseudomonas aeruginosa PAO1 | | Descriptor: | hypothetical protein PA1206 | | Authors: | Zimmerman, M.D, Chruszcz, M, Cymborowski, M.T, Wang, S, Kirillova, O, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-20 | | Release date: | 2006-01-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of PR10-Allergen-Like Protein PA1206 From Pseudomonas aeruginosa PAO1
To be Published
|
|
3VDH
 
 | | Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14 | | Descriptor: | B-1,4-endoglucanase, CHLORIDE ION | | Authors: | Stogios, P.J, Evdokimova, E, Egorova, O, Yim, V, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-01-05 | | Release date: | 2012-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-Function Analysis of a Mixed-linkage beta-Glucanase/Xyloglucanase from the Key Ruminal Bacteroidetes Prevotella bryantii B14.
J.Biol.Chem., 291, 2016
|
|
