4Z8M
 
 | | Crystal structure of the MAVS-TRAF6 complex | | Descriptor: | Peptide from Mitochondrial antiviral-signaling protein, TNF receptor-associated factor 6 | | Authors: | Shi, Z.B, Zhou, Z. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Insights into Mitochondrial Antiviral Signaling Protein (MAVS)-Tumor Necrosis Factor Receptor-associated Factor 6 (TRAF6) Signaling
J.Biol.Chem., 290, 2015
|
|
2I3R
 
 | | Engineered catalytic domain of protein tyrosine phosphatase HPTPbeta | | Descriptor: | CHLORIDE ION, Receptor-type tyrosine-protein phosphatase beta | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-08-20 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering the catalytic domain of human protein tyrosine phosphatase beta for structure-based drug discovery.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2I3U
 
 | | Structural studies of protein tyrosine phosphatase beta catalytic domain in complex with inhibitors | | Descriptor: | Receptor-type tyrosine-protein phosphatase beta | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-08-20 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering the catalytic domain of human protein tyrosine phosphatase beta for structure-based drug discovery.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2I4E
 
 | | Structural studies of protein tyrosine phosphatase beta catalytic domain in complex with inhibitors | | Descriptor: | Receptor-type tyrosine-protein phosphatase beta, VANADATE ION | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-08-21 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineering the catalytic domain of human protein tyrosine phosphatase beta for structure-based drug discovery.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2I4H
 
 | | Structural studies of protein tyrosine phosphatase beta catalytic domain co-crystallized with a sulfamic acid inhibitor | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-(TERT-BUTOXYCARBONYL)-L-TYROSYL-N-METHYL-4-(SULFOAMINO)-L-PHENYLALANINAMIDE, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-08-21 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Engineering the catalytic domain of human protein tyrosine phosphatase beta for structure-based drug discovery.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
6MUL
 
 | | Murine PI3K delta kinsae domain - cpd 1 | | Descriptor: | 1-{1-[8-(1-ethyl-5-methyl-1H-pyrazol-4-yl)-9-methyl-9H-purin-6-yl]piperidin-4-yl}-1,3-dihydro-2H-imidazo[4,5-b]pyridin-2-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Fischmann, T.O. | | Deposit date: | 2018-10-23 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structure Overhaul Affords a Potent Purine PI3K delta Inhibitor with Improved Tolerability.
J.Med.Chem., 62, 2019
|
|
6MUM
 
 | | Murine PI3K delta kinsae domain - cpd 3 | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, [(3S)-3-{[8-(1-ethyl-5-methyl-1H-pyrazol-4-yl)-9-methyl-9H-purin-6-yl]oxy}pyrrolidin-1-yl](oxan-4-yl)methanone | | Authors: | Fischmann, T.O. | | Deposit date: | 2018-10-23 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structure Overhaul Affords a Potent Purine PI3K delta Inhibitor with Improved Tolerability.
J.Med.Chem., 62, 2019
|
|
1JTF
 
 | | Crystal Structure Analysis of VP39-F180W mutant and m7GpppG complex | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Oguro, A, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 2001-08-20 | | Release date: | 2002-07-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The "cap-binding slot" of an mRNA cap-binding protein: quantitative effects of aromatic side chain choice in the double-stacking sandwich with cap.
Biochemistry, 41, 2002
|
|
6NCJ
 
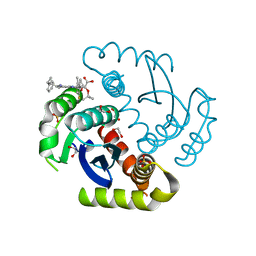 | | Structure of HIV-1 Integrase with potent 5,6,7,8-Tetrahydro-1,6-naphthyridine Derivatives Allosteric Site Inhibitors | | Descriptor: | (2~{S})-2-[4-(8-fluoranyl-5-methyl-3,4-dihydro-2~{H}-chromen-6-yl)-2-methyl-6-[[(1~{S},2~{R})-2-phenylcyclopropyl]methyl]-7,8-dihydro-5~{H}-1,6-naphthyridin-3-yl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, 1,2-ETHANEDIOL, Integrase, ... | | Authors: | Nolte, R.T. | | Deposit date: | 2018-12-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 5,6,7,8-Tetrahydro-1,6-naphthyridine Derivatives as Potent HIV-1-Integrase-Allosteric-Site Inhibitors.
J. Med. Chem., 62, 2019
|
|
6EI3
 
 | | Crystal structure of auto inhibited POT family peptide transporter | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Proton-dependent oligopeptide transporter family protein | | Authors: | Newstead, S, Brinth, A, Vogeley, L, Caffrey, M. | | Deposit date: | 2017-09-17 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Proton movement and coupling in the POT family of peptide transporters.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1JSZ
 
 | | Crystal Structure Analysis of N7,9-dimethylguanine-VP39 complex | | Descriptor: | 7,9-DIMETHYLGUANINE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Oguro, A, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 2001-08-19 | | Release date: | 2002-07-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The "cap-binding slot" of an mRNA cap-binding protein: quantitative effects of aromatic side chain choice in the double-stacking sandwich with cap.
Biochemistry, 41, 2002
|
|
1JTE
 
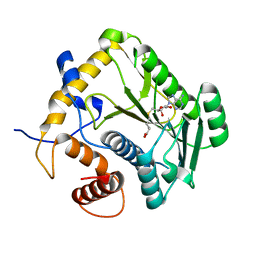 | | Crystal Structure Analysis of VP39 F180W mutant | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Oguro, A, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 2001-08-20 | | Release date: | 2002-07-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The "cap-binding slot" of an mRNA cap-binding protein: quantitative effects of aromatic side chain choice in the double-stacking sandwich with cap.
Biochemistry, 41, 2002
|
|
6DQG
 
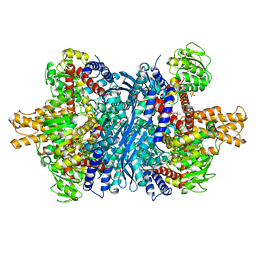 | | Human glutamate dehydrogenase, H454Y mutant | | Descriptor: | Glutamate dehydrogenase 1, mitochondrial, PHOSPHATE ION | | Authors: | Smith, T.J. | | Deposit date: | 2018-06-10 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Glutamate dehydrogenase: Structure of a hyperinsulinism mutant, corrections to the atomic model, and insights into a regulatory site.
Proteins, 87, 2019
|
|
3CQW
 
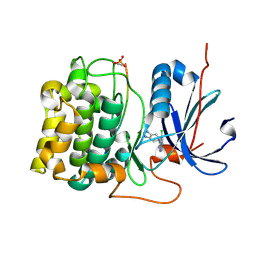 | | Crystal Structure of Akt-1 complexed with substrate peptide and inhibitor | | Descriptor: | 5-(5-chloro-7H-pyrrolo[2,3-d]pyrimidin-4-yl)-4,5,6,7-tetrahydro-1H-imidazo[4,5-c]pyridine, Glycogen synthase kinase-3 beta, MANGANESE (II) ION, ... | | Authors: | Pandit, J. | | Deposit date: | 2008-04-03 | | Release date: | 2008-05-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and structure based optimization of novel Akt inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6TCV
 
 | | Crystal structure of Bacteroides thetaiotamicron EndoBT-3987 in complex with Man9GlcNAc2Asn substrate | | Descriptor: | ASPARAGINE, Endo-beta-N-acetylglucosaminidase F1, GLYCEROL, ... | | Authors: | Trastoy, B, Du, J.J, Klontz, E.H, Cifuente, J.O, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2019-11-06 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.311 Å) | | Cite: | Structural basis of mammalian high-mannose N-glycan processing by human gut Bacteroides.
Nat Commun, 11, 2020
|
|
6T8K
 
 | | Crystal structure of Bacteroides thetaiotamicron EndoBT-3987 in complex with Man9GlcNAc product in P1 | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase F1, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Trastoy, B, Du, J.J, Klontz, E.H, Cifuente, J.O, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2019-10-24 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of mammalian high-mannose N-glycan processing by human gut Bacteroides.
Nat Commun, 11, 2020
|
|
6T8I
 
 | | Crystal structure of wild type EndoBT-3987 from Bacteroides thetaiotamicron VPI-5482 | | Descriptor: | Endo-beta-N-acetylglucosaminidase F1, GLYCEROL | | Authors: | Trastoy, B, Du, J.J, Klontz, E.H, Cifuente, J.O, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2019-10-24 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of mammalian high-mannose N-glycan processing by human gut Bacteroides.
Nat Commun, 11, 2020
|
|
6T8L
 
 | | Crystal structure of Bacteroides thetaiotamicron EndoBT-3987 with Man9GlcNAc product in P212121 | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase F1, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Trastoy, B, Du, J.J, Klontz, E.H, Cifuente, J.O, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2019-10-24 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of mammalian high-mannose N-glycan processing by human gut Bacteroides.
Nat Commun, 11, 2020
|
|
6TCW
 
 | | Crystal structure of Bacteroides thetaiotamicron EndoBT-3987 with Man5GlcNAc product | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase F1, GLYCEROL, ... | | Authors: | Trastoy, B, Du, J.J, Klontz, E.H, Cifuente, J.O, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2019-11-06 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Structural basis of mammalian high-mannose N-glycan processing by human gut Bacteroides.
Nat Commun, 11, 2020
|
|
7X7W
 
 | | The X-ray Crystallographic Structure of D-Psicose 3-epimerase from Clostridia bacterium | | Descriptor: | D-PSICOSE 3-EPIMERASE, MANGANESE (II) ION | | Authors: | Li, Z.F, Ban, X.F, Xie, X.F, Tian, Y.X, Li, C.M, Gu, Z.B. | | Deposit date: | 2022-03-10 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Crystal structure of a novel homodimeric D-allulose 3-epimerase from a Clostridia bacterium
Acta Crystallogr.,Sect.D, 78, 2022
|
|
5J9U
 
 | | Crystal structure of the NuA4 core complex | | Descriptor: | Chromatin modification-related protein EAF6, Chromatin modification-related protein YNG2, Enhancer of polycomb-like protein 1, ... | | Authors: | Chen, Z.C, Xu, P. | | Deposit date: | 2016-04-11 | | Release date: | 2016-10-26 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The NuA4 Core Complex Acetylates Nucleosomal Histone H4 through a Double Recognition Mechanism
Mol.Cell, 63, 2016
|
|
6L8Q
 
 | | Complex structure of bat CD26 and MERS-RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Yuan, Y. | | Deposit date: | 2019-11-07 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular Basis of Binding between Middle East Respiratory Syndrome Coronavirus and CD26 from Seven Bat Species.
J.Virol., 94, 2020
|
|
5J9W
 
 | | Crystal structure of the NuA4 core complex | | Descriptor: | ACETYL COENZYME *A, Chromatin modification-related protein EAF6, Chromatin modification-related protein YNG2, ... | | Authors: | Chen, Z.C, Xu, P. | | Deposit date: | 2016-04-11 | | Release date: | 2016-10-26 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The NuA4 Core Complex Acetylates Nucleosomal Histone H4 through a Double Recognition Mechanism
Mol.Cell, 63, 2016
|
|
5J9T
 
 | | Crystal structure of the NuA4 core complex | | Descriptor: | Chromatin modification-related protein EAF6, Chromatin modification-related protein YNG2, Enhancer of polycomb-like protein 1, ... | | Authors: | Chen, Z.C, Xu, P. | | Deposit date: | 2016-04-11 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The NuA4 Core Complex Acetylates Nucleosomal Histone H4 through a Double Recognition Mechanism
Mol.Cell, 63, 2016
|
|
6M4G
 
 | | Structural mechanism of nucleosome dynamics governed by human histone variants H2A.B and H2A.Z.2.2 | | Descriptor: | DNA (93-MER), Histone H2A-Bbd type 2/3, Histone H2B type 2-E, ... | | Authors: | Zhou, M, Dai, L.C, Li, C.M, Shi, L.X, Huang, Y, Guo, Z.Q. | | Deposit date: | 2020-03-06 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of nucleosome dynamics modulation by histone variants H2A.B and H2A.Z.2.2.
Embo J., 40, 2021
|
|
