4K3J
 
 | | Crystal structure of Onartuzumab Fab in complex with MET and HGF-beta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor beta chain, ... | | Authors: | Ma, X, Starovasnik, M.A. | | Deposit date: | 2013-04-10 | | Release date: | 2013-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Monovalent antibody design and mechanism of action of onartuzumab, a MET antagonist with anti-tumor activity as a therapeutic agent.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1SHO
 
 | |
4W9Y
 
 | |
7O3W
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | Authors: | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
7O3X
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | Authors: | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
7O40
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | Authors: | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
7O3Y
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | Authors: | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
7O3Z
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | Authors: | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
8CJ9
 
 | | Crystal structure of maize CKO/CKX8 in complex with urea-derived inhibitor 2-[(3,5-dichlorophenyl)carbamoylamino]benzamide | | Descriptor: | 1,2-ETHANEDIOL, 2-[[3,5-bis(chloranyl)phenyl]carbamoylamino]benzamide, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2023-02-12 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Cytokinin oxidase/dehydrogenase inhibitors: progress towards agricultural practice.
J.Exp.Bot., 75, 2024
|
|
7O49
 
 | |
7O4A
 
 | |
7O4C
 
 | |
6Z0F
 
 | | Crystal structure of the membrane pseudokinase YukC/EssB from Bacillus subtilis T7SS | | Descriptor: | ESX secretion system protein YukC | | Authors: | Tassinari, M, Bellinzoni, M, Alzari, P.M, Fronzes, R, Gubellini, F. | | Deposit date: | 2020-05-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | The Antibacterial Type VII Secretion System of Bacillus subtilis: Structure and Interactions of the Pseudokinase YukC/EssB.
Mbio, 13, 2022
|
|
5CAL
 
 | | EGFR kinase domain mutant "TMLR" with compound 24 | | Descriptor: | 2,2-dimethyl-3-[(4-{[2-methyl-1-(propan-2-yl)-1H-imidazo[4,5-c]pyridin-6-yl]amino}pyrimidin-2-yl)amino]propanamide, Epidermal growth factor receptor | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2015-06-29 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Noncovalent Mutant Selective Epidermal Growth Factor Receptor Inhibitors: A Lead Optimization Case Study.
J.Med.Chem., 58, 2015
|
|
5C8N
 
 | | EGFR kinase domain mutant "TMLR" with compound 23 | | Descriptor: | Epidermal growth factor receptor, N-{2-[4-(2-aminoethyl)-4-methoxypiperidin-1-yl]pyrimidin-4-yl}-2-methyl-1-(propan-2-yl)-1H-imidazo[4,5-c]pyridin-6-amine, SULFATE ION | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2015-06-25 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Noncovalent Mutant Selective Epidermal Growth Factor Receptor Inhibitors: A Lead Optimization Case Study.
J.Med.Chem., 58, 2015
|
|
5C8K
 
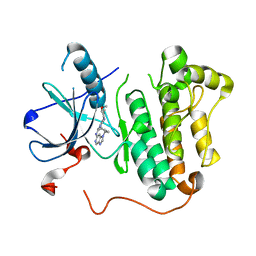 | | EGFR kinase domain mutant "TMLR" with compound 1 | | Descriptor: | 1-cyclopentyl-N-[2-(4-methoxypiperidin-1-yl)pyrimidin-4-yl]-1H-imidazo[4,5-c]pyridin-6-amine, Epidermal growth factor receptor | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2015-06-25 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Noncovalent Mutant Selective Epidermal Growth Factor Receptor Inhibitors: A Lead Optimization Case Study.
J.Med.Chem., 58, 2015
|
|
5CAP
 
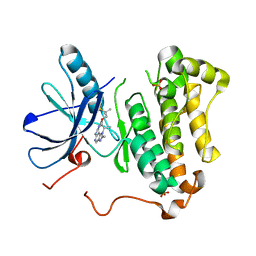 | | EGFR kinase domain mutant "TMLR" with compound 30 | | Descriptor: | 2-methyl-N-[2-(2-methyl-2-methylsulfonyl-propoxy)pyrimidin-4-yl]-1-propan-2-yl-imidazo[4,5-c]pyridin-6-amine, Epidermal growth factor receptor, SULFATE ION | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2015-06-29 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Noncovalent Mutant Selective Epidermal Growth Factor Receptor Inhibitors: A Lead Optimization Case Study.
J.Med.Chem., 58, 2015
|
|
2P7M
 
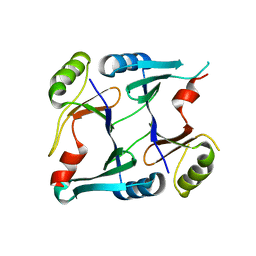 | | Crystal structure of monoclinic form of genomically encoded fosfomycin resistance protein, FosX, from Listeria monocytogenes at pH 6.5 | | Descriptor: | Glyoxalase family protein | | Authors: | Fillgrove, K.L, Pakhomova, S, Schaab, M, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2007-03-20 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Mechanism of the Genomically Encoded Fosfomycin Resistance Protein, FosX, from Listeria monocytogenes.
Biochemistry, 46, 2007
|
|
2P7L
 
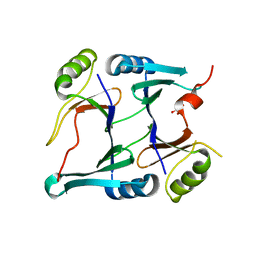 | | Crystal structure of monoclinic form of genomically encoded fosfomycin resistance protein, FosX, from Listeria monocytogenes at pH 5.75 | | Descriptor: | GLYCEROL, Glyoxalase family protein | | Authors: | Fillgrove, K.L, Pakhomova, S, Schaab, M, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2007-03-20 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Mechanism of the Genomically Encoded Fosfomycin Resistance Protein, FosX, from Listeria monocytogenes.
Biochemistry, 46, 2007
|
|
2P7P
 
 | | Crystal structure of genomically encoded fosfomycin resistance protein, FosX, from Listeria monocytogenes complexed with MN(II) and sulfate ion | | Descriptor: | Glyoxalase family protein, MANGANESE (II) ION, SULFATE ION | | Authors: | Fillgrove, K.L, Pakhomova, S, Schaab, M, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2007-03-20 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure and Mechanism of the Genomically Encoded Fosfomycin Resistance Protein, FosX, from Listeria monocytogenes.
Biochemistry, 46, 2007
|
|
8CD9
 
 | | Cathepsin B1 from Schistosoma mansoni in complex with gallinamide analog 6 | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin B-like peptidase (C01 family), SODIUM ION, ... | | Authors: | Rubesova, P, Brynda, J, Fanfrlik, J, Gerwick, W.H, Mares, M. | | Deposit date: | 2023-01-30 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Nature-Inspired Gallinamides Are Potent Antischistosomal Agents: Inhibition of the Cathepsin B1 Protease Target and Binding Mode Analysis.
Acs Infect Dis., 10, 2024
|
|
8C8P
 
 | |
8QJR
 
 | | BRG1 bromodomain in complex with VBC via compound 17 | | Descriptor: | (2S,4R)-1-[(2R)-2-[3-[2-[4-[3-[4-[(1R,5S)-3-[3-azanyl-6-(2-hydroxyphenyl)pyridazin-4-yl]-3,8-diazabicyclo[3.2.1]octan-8-yl]pyridin-2-yl]oxycyclobutyl]oxypiperidin-1-yl]ethoxy]-1,2-oxazol-5-yl]-3-methyl-butanoyl]-N-[(1S)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, CHLORIDE ION, Elongin-B, ... | | Authors: | Kerry, P.S, Hole, A.J, Perez-Dorado, J.I. | | Deposit date: | 2023-09-13 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | PROTACs Targeting BRM (SMARCA2) Afford Selective In Vivo Degradation over BRG1 (SMARCA4) and Are Active in BRG1 Mutant Xenograft Tumor Models.
J.Med.Chem., 67, 2024
|
|
8QJS
 
 | | VHL/Elongin B/Elongin C complex with compound 155 | | Descriptor: | (2S,4R)-1-[(2R)-2-[3-[2-(2-methoxyethoxy)ethoxy]-1,2-oxazol-5-yl]-3-methyl-butanoyl]-N-[(1S)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Kerry, P.S, Hole, A.J, Perez-Dorado, J.I. | | Deposit date: | 2023-09-13 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.191 Å) | | Cite: | PROTACs Targeting BRM (SMARCA2) Afford Selective In Vivo Degradation over BRG1 (SMARCA4) and Are Active in BRG1 Mutant Xenograft Tumor Models.
J.Med.Chem., 67, 2024
|
|
8QJT
 
 | | BRM (SMARCA2) Bromodomain in complex with ligand 10 | | Descriptor: | 2-[6-azanyl-5-[(1R,5S)-8-[2-(2-methoxyethoxy)pyridin-4-yl]-3,8-diazabicyclo[3.2.1]octan-3-yl]pyridazin-3-yl]phenol, CHLORIDE ION, Probable global transcription activator SNF2L2, ... | | Authors: | Kerry, P.S, Hole, A.J, Perez-Dorado, J.I. | | Deposit date: | 2023-09-13 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.568 Å) | | Cite: | PROTACs Targeting BRM (SMARCA2) Afford Selective In Vivo Degradation over BRG1 (SMARCA4) and Are Active in BRG1 Mutant Xenograft Tumor Models.
J.Med.Chem., 67, 2024
|
|
