4G08
 
 | |
7JZN
 
 | |
4R6G
 
 | |
7JZL
 
 | |
5BYO
 
 | |
4EEF
 
 | |
7JZM
 
 | |
4PEK
 
 | | Crystal structure of a computationally designed retro-aldolase, RA114.3 | | Descriptor: | Retro-aldolase | | Authors: | Bhabha, G, Zhang, X, Liu, Y, Ekiert, D.C. | | Deposit date: | 2014-04-23 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | De novo-designed enzymes as small-molecule-regulated fluorescence imaging tags and fluorescent reporters.
J.Am.Chem.Soc., 136, 2014
|
|
4PEJ
 
 | | Crystal structure of a computationally designed retro-aldolase, RA110.4 (Cys free) | | Descriptor: | Retro-aldolase | | Authors: | Bhabha, G, Zhang, X, Liu, Y, Ekiert, D.C. | | Deposit date: | 2014-04-23 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | De novo-designed enzymes as small-molecule-regulated fluorescence imaging tags and fluorescent reporters.
J.Am.Chem.Soc., 136, 2014
|
|
5BVB
 
 | | Engineered Digoxigenin binder DIG5.1a | | Descriptor: | DIG5.1a, DIGOXIGENIN | | Authors: | Doyle, L.A, Stoddard, B.L. | | Deposit date: | 2015-06-04 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | CSAR Benchmark Exercise 2013: Evaluation of Results from a Combined Computational Protein Design, Docking, and Scoring/Ranking Challenge.
J.Chem.Inf.Model., 56, 2016
|
|
7JZU
 
 | |
5BVL
 
 | |
4G2S
 
 | |
5CWF
 
 | |
4R58
 
 | |
4R5C
 
 | |
4QQ8
 
 | | Crystal structure of the formolase FLS in space group P 43 21 2 | | Descriptor: | 1,2-ETHANEDIOL, Formolase, MAGNESIUM ION, ... | | Authors: | Shen, B.W, Siegel, J.B, Stoddard, B.L. | | Deposit date: | 2014-06-26 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Computational protein design enables a novel one-carbon assimilation pathway.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8UOS
 
 | |
8UPB
 
 | |
8UPA
 
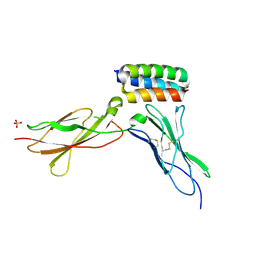 | | Structure of gp130 in complex with a de novo designed IL-6 mimetic | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, De novo designed IL-6 mimetic, Interleukin-6 receptor subunit beta, ... | | Authors: | Borowska, M.T, Jude, K.M, Garcia, K.C. | | Deposit date: | 2023-10-22 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | De novo design of miniprotein antagonists of cytokine storm inducers.
Nat Commun, 15, 2024
|
|
7YH8
 
 | | Crystal structure of a heterochiral protein complex | | Descriptor: | D-Pep-1, L-19437 | | Authors: | Liang, M, Li, S, Wang, T, Liu, L, Lu, P. | | Deposit date: | 2022-07-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Accurate de novo design of heterochiral protein-protein interactions
Cell Res., 2024
|
|
4OYC
 
 | |
4OU1
 
 | | Crystal structure of a computationally designed retro-aldolase covalently bound to folding probe 1 [(6-methoxynaphthalen-2-yl)(oxiran-2-yl)methanol] | | Descriptor: | (1S,2S)-1-(6-methoxynaphthalen-2-yl)propane-1,2-diol, BENZOIC ACID, PHOSPHATE ION, ... | | Authors: | Bhabha, G, Zhang, X, Ekiert, D.C. | | Deposit date: | 2014-02-14 | | Release date: | 2014-03-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Small molecule probes to quantify the functional fraction of a specific protein in a cell with minimal folding equilibrium shifts.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4N9G
 
 | |
3U43
 
 | |
