7M3X
 
 | | Crystal Structure of the Apo Form of Human RBBP7 | | Descriptor: | Histone-binding protein RBBP7, UNKNOWN ATOM OR ION | | Authors: | Righetto, G.L, Dong, A, Li, Y, Hutchinson, A, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-03-19 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal Structure of the Apo Form of Human RBBP7
To Be Published
|
|
8HDK
 
 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine and glutamate (minor class in symmetry) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-11-04 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7VCT
 
 | | Human p97 single hexamer conformer III with D1-ATPgammaS and D2-ADP bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gao, H, Li, F, Shi, Z, Li, Y, Yu, H. | | Deposit date: | 2021-09-04 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM structures of human p97 double hexamer capture potentiated ATPase-competent state.
Cell Discov, 8, 2022
|
|
7VCX
 
 | | Human p97 single hexamer conformer II with ATPgammaS bound | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Gao, H, Li, F, Shi, Z, Li, Y, Yu, H. | | Deposit date: | 2021-09-04 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Cryo-EM structures of human p97 double hexamer capture potentiated ATPase-competent state.
Cell Discov, 8, 2022
|
|
7VCS
 
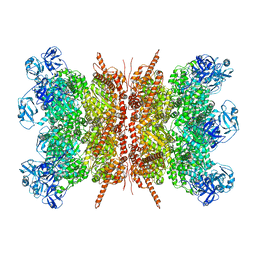 | | Human p97 double hexamer conformer II with ATPgammaS bound | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Gao, H, Li, F, Shi, Z, Li, Y, Yu, H. | | Deposit date: | 2021-09-03 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-EM structures of human p97 double hexamer capture potentiated ATPase-competent state.
Cell Discov, 8, 2022
|
|
7VCV
 
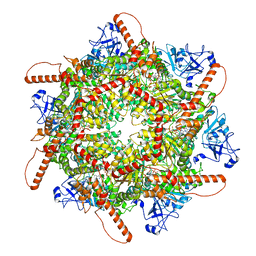 | | Human p97 single hexamer conformer I with ATPgammaS bound | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Gao, H, Li, F, Shi, Z, Li, Y, Yu, H. | | Deposit date: | 2021-09-04 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM structures of human p97 double hexamer capture potentiated ATPase-competent state.
Cell Discov, 8, 2022
|
|
7MIR
 
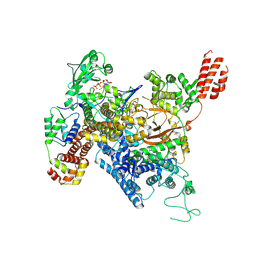 | | Cryo-EM structure of SidJ-SdeA-CaM reaction intermediate complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Osinski, A, Black, M.H, Pawlowski, K, Chen, Z, Li, Y, Tagliabracci, V.S. | | Deposit date: | 2021-04-17 | | Release date: | 2021-08-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural and mechanistic basis for protein glutamylation by the kinase fold.
Mol.Cell, 81, 2021
|
|
7MIS
 
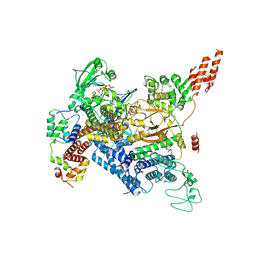 | | Cryo-EM structure of SidJ-SdeC-CaM reaction intermediate complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Osinski, A, Black, M.H, Pawlowski, K, Chen, Z, Li, Y, Tagliabracci, V.S. | | Deposit date: | 2021-04-17 | | Release date: | 2021-08-18 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural and mechanistic basis for protein glutamylation by the kinase fold.
Mol.Cell, 81, 2021
|
|
4FGY
 
 | | Identification of a unique PPAR ligand with an unexpected binding mode and antibetic activity | | Descriptor: | (4R,6S,8S,12R,14R,16Z,18R,19R,20S,21S)-19,21-dihydroxy-22-{(2S,2'R,5S,5'S)-5'-[(1R)-1-hydroxyethyl]-2,5'-dimethyloctahydro-2,2'-bifuran-5-yl}-4,6,8,12,14,18,20-heptamethyl-9,11-dioxodocos-16-enoic acid, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Wang, R, Li, Y. | | Deposit date: | 2012-06-05 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Identification of the antibiotic ionomycin as an unexpected peroxisome proliferator-activated receptor Gamma (PPAR-gamma) ligand with a unique binding mode and effective glucose-lowering activity in a mouse model of diabetes.
Diabetologia, 56, 2013
|
|
3HD2
 
 | | Crystal structure of E. coli HPPK(Q50A) in complex with MgAMPCPP and pterin | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-05-06 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Role of loop coupling in enzymatic catalysis and conformational dynamics
To be Published
|
|
3HD1
 
 | | Crystal structure of E. coli HPPK(N10A) in complex with MgAMPCPP | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-05-06 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Role of loop coupling in enzymatic catalysis and conformational dynamics
To be Published
|
|
1X9M
 
 | | T7 DNA polymerase in complex with an N-2-acetylaminofluorene-adducted DNA | | Descriptor: | 5'-D(*CP*CP*CP*(8FG)P*AP*TP*CP*AP*CP*AP*CP*TP*AP*CP*CP*AP*AP*TP*CP*AP*CP*TP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*AP*GP*TP*GP*AP*TP*TP*GP*GP*TP*AP*GP*TP*GP*TP*GP*AP*(2DT))-3', DNA polymerase, ... | | Authors: | Dutta, S, Li, Y, Johnson, D, Dzantiev, L, Richardson, C.C, Romano, L.J, Ellenberger, T. | | Deposit date: | 2004-08-23 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of 2-acetylaminofluorene and 2-aminofluorene in complex with T7 DNA polymerase reveal mechanisms of mutagenesis.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1X9W
 
 | | T7 DNA polymerase in complex with a primer/template DNA containing a disordered N-2 aminofluorene on the template, crystallized with dideoxy-ATP as the incoming nucleotide. | | Descriptor: | 5'-D(*CP*CP*CP*(AFG)*AP*TP*CP*AP*CP*AP*CP*TP*AP*CP*CP*AP*AP*TP*CP*AP*CP*TP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*AP*GP*TP*GP*AP*TP*T*GP*GP*T*AP*GP*TP*GP*TP*GP*AP*(2DT))-3', DNA polymerase, ... | | Authors: | Dutta, S, Li, Y, Johnson, D, Dzantiev, L, Richardson, C.C, Romano, L.J, Ellenberger, T. | | Deposit date: | 2004-08-24 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of 2-acetylaminofluorene and 2-aminofluorene in complex with T7 DNA polymerase reveal mechanisms of mutagenesis.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1YNF
 
 | | Crystal Structure of N-Succinylarginine Dihydrolase, AstB, bound to Substrate and Product, an Enzyme from the Arginine Catabolic Pathway of Escherichia coli | | Descriptor: | POTASSIUM ION, Succinylarginine dihydrolase | | Authors: | Tocilj, A, Schrag, J.D, Li, Y, Schneider, B.L, Reitzer, L, Matte, A, Cygler, M. | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of N-succinylarginine dihydrolase AstB, bound to substrate and product, an enzyme from the arginine catabolic pathway of Escherichia coli.
J.Biol.Chem., 280, 2005
|
|
5ZUE
 
 | | GTP-bound, double-stranded, curved FtsZ protofilament structure | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Guan, F, Yu, J, Yu, J, Liu, Y, Li, Y, Feng, X.H, Huang, K.C, Chang, Z, Ye, S. | | Deposit date: | 2018-05-07 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Lateral interactions between protofilaments of the bacterial tubulin homolog FtsZ are essential for cell division
Elife, 7, 2018
|
|
3O73
 
 | | Crystal structure of quinone reductase 2 in complex with the indolequinone MAC627 | | Descriptor: | 5-[(4-aminobutyl)amino]-1,2-dimethyl-3-[(4-nitrophenoxy)methyl]-1H-indole-4,7-dione, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Dufour, M, Yan, C, Colucci, M.A, Siegel, D, Li, Y, De Matteis, C.I, Ross, D, Moody, C.J. | | Deposit date: | 2010-07-30 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism-Based Inhibition of Quinone Reductase 2 (NQO2): Selectivity for NQO2 over NQO1 and Structural Basis for Flavoprotein Inhibition.
Chembiochem, 12, 2011
|
|
3IV1
 
 | | Coiled-coil domain of tumor susceptibility gene 101 | | Descriptor: | CHLORIDE ION, SULFATE ION, Tumor susceptibility gene 101 protein | | Authors: | Neculai, D, Avvakumov, G.V, Wernimont, A.K, Xue, S, Walker, J.R, Li, Y, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-31 | | Release date: | 2009-11-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Coiled-Coil Domain of Human Tsg101
To be Published
|
|
8GCY
 
 | | Co-crystal structure of CBL-B in complex with N-Aryl isoindolin-1-one inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-{3-[(1s,3R)-3-methyl-1-(4-methyl-4H-1,2,4-triazol-3-yl)cyclobutyl]phenyl}-6-{[(3S)-3-methylpiperidin-1-yl]methyl}-4-(trifluoromethyl)-2,3-dihydro-1H-isoindol-1-one, E3 ubiquitin-protein ligase CBL-B, ... | | Authors: | Kimani, S, Zeng, H, Dong, A, Li, Y, Santhakumar, V, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-03 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The co-crystal structure of Cbl-b and a small-molecule inhibitor reveals the mechanism of Cbl-b inhibition.
Commun Biol, 6, 2023
|
|
1YNI
 
 | | Crystal Structure of N-Succinylarginine Dihydrolase, AstB, bound to Substrate and Product, an Enzyme from the Arginine Catabolic Pathway of Escherichia coli | | Descriptor: | N~2~-(3-CARBOXYPROPANOYL)-L-ARGININE, POTASSIUM ION, Succinylarginine Dihydrolase | | Authors: | Tocilj, A, Schrag, J.D, Li, Y, Schneider, B.L, Reitzer, L, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of N-succinylarginine dihydrolase AstB, bound to substrate and product, an enzyme from the arginine catabolic pathway of Escherichia coli.
J.Biol.Chem., 280, 2005
|
|
1YNH
 
 | | Crystal Structure of N-Succinylarginine Dihydrolase, AstB, bound to Substrate and Product, an Enzyme from the Arginine Catabolic Pathway of Escherichia coli | | Descriptor: | N~2~-(3-CARBOXYPROPANOYL)-L-ORNITHINE, POTASSIUM ION, Succinylarginine Dihydrolase | | Authors: | Tocilj, A, Schrag, J.D, Li, Y, Schneider, B.L, Reitzer, L, Matte, A, Cygler, M. | | Deposit date: | 2005-01-24 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of N-succinylarginine dihydrolase AstB, bound to substrate and product, an enzyme from the arginine catabolic pathway of Escherichia coli.
J.Biol.Chem., 280, 2005
|
|
8T42
 
 | | Model of TTLL6 MTBH1-2 bound to microtubule | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Mahalingan, K.K, Grotjahn, D, Li, Y, Lander, G.C, Zehr, E.A, Roll-Mecak, A. | | Deposit date: | 2023-06-08 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for alpha-tubulin-specific and modification state-dependent glutamylation.
Nat.Chem.Biol., 20, 2024
|
|
1XC5
 
 | | Solution Structure of the SMRT Deacetylase Activation Domain | | Descriptor: | Nuclear receptor corepressor 2 | | Authors: | Codina, A, Love, J.D, Li, Y, Lazar, M.A, Neuhaus, D, Schwabe, J.W.R. | | Deposit date: | 2004-09-01 | | Release date: | 2005-05-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the interaction and activation of histone deacetylase 3 by nuclear receptor corepressors
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3JA9
 
 | |
3JAA
 
 | | HUMAN DNA POLYMERASE ETA in COMPLEX WITH NORMAL DNA AND INCO NUCLEOTIDE (NRM) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*T*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*T)-3, DNA (5'-D(*TP*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | Authors: | Lau, W.C.Y, Li, Y, Zhang, Q, Huen, M.S.Y. | | Deposit date: | 2015-05-19 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (22 Å) | | Cite: | Molecular architecture of the Ub-PCNA/Pol eta complex bound to DNA
Sci Rep, 5, 2015
|
|
1YVJ
 
 | | Crystal structure of the Jak3 kinase domain in complex with a staurosporine analogue | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 1,2,3,4-TETRAHYDROGEN-STAUROSPORINE, Tyrosine-protein kinase JAK3 | | Authors: | Boggon, T.J, Li, Y, Manley, P.W, Eck, M.J. | | Deposit date: | 2005-02-15 | | Release date: | 2005-05-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the Jak3 kinase domain in complex with a staurosporine analog
Blood, 106, 2005
|
|
