7T3J
 
 | | Cryo-EM structure of Csy-AcrIF24 | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR type I-F/YPEST-associated protein Csy3, ... | | Authors: | Mukherjee, I.A, Chang, L. | | Deposit date: | 2021-12-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of AcrIF24 as an anti-CRISPR protein and transcriptional suppressor.
Nat.Chem.Biol., 18, 2022
|
|
7TAW
 
 | | Cryo-EM structure of the Csy-AcrIF24-promoter DNA dimer | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR type I-F/YPEST-associated protein Csy3, ... | | Authors: | Mukherjee, I.A, Chang, L. | | Deposit date: | 2021-12-21 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of AcrIF24 as an anti-CRISPR protein and transcriptional suppressor.
Nat.Chem.Biol., 18, 2022
|
|
7TAX
 
 | | Cryo-EM structure of the Csy-AcrIF24-promoter DNA complex | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR type I-F/YPEST-associated protein Csy3, ... | | Authors: | Mukherjee, I.A, Chang, L. | | Deposit date: | 2021-12-21 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of AcrIF24 as an anti-CRISPR protein and transcriptional suppressor.
Nat.Chem.Biol., 18, 2022
|
|
7T3L
 
 | | Cryo-EM structure of Csy-AcrIF24-DNA dimer | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR type I-F/YPEST-associated protein Csy3, ... | | Authors: | Mukherjee, I.A, Chang, L. | | Deposit date: | 2021-12-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of AcrIF24 as an anti-CRISPR protein and transcriptional suppressor.
Nat.Chem.Biol., 18, 2022
|
|
4KTL
 
 | | Crystal structure of Mycobacterium tuberculosis CYP121 in complex with 4,4'-(3-((4-hydroxyphenyl)amino)-1H-pyrazole-4,5-diyl)diphenol | | Descriptor: | 4,4'-{3-[(4-hydroxyphenyl)amino]-1H-pyrazole-4,5-diyl}diphenol, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Hudson, S.A. | | Deposit date: | 2013-05-20 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Overcoming the Limitations of Fragment Merging: Rescuing a Strained Merged Fragment Series Targeting Mycobacterium tuberculosis CYP121.
Chemmedchem, 8, 2013
|
|
4KTJ
 
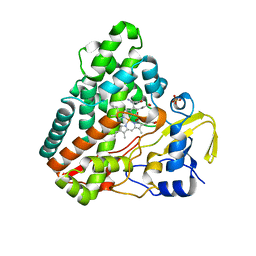 | |
4KTK
 
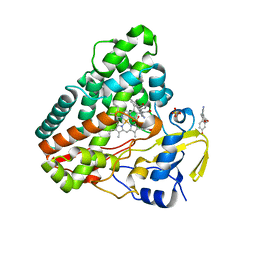 | |
7QZX
 
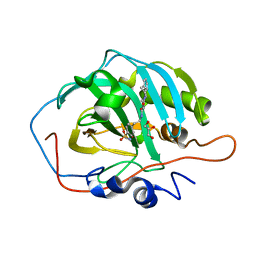 | | Human Carbonic Anhydrase II in complex with 4-oxo-N-(4-sulfamoylphenethyl)-1,3,4,6,7,11b-hexahydro-2H-pyrazino[2,1-a]isoquinoline-2-carboxamide | | Descriptor: | 4-oxo-N-(4-sulfamoylphenethyl)-1,3,4,6,7,11b-hexahydro-2H-pyrazino[2,1-a]isoquinoline-2-carboxamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2022-02-01 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Development of Praziquantel sulphonamide derivatives as antischistosomal drugs.
J Enzyme Inhib Med Chem, 37, 2022
|
|
7R1X
 
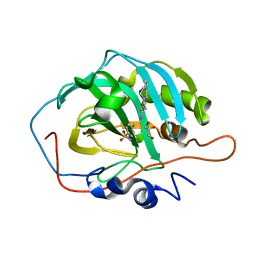 | | human Carbonic Anhydrase II in complex with 4-oxo-N-(4-sulfamoylphenethyl)-1,3,4,6,7,11b-hexahydro-2H-pyrazino[2,1-a]isoquinoline-2-carbothioamide | | Descriptor: | 4-oxo-N-(4-sulfamoylphenethyl)-1,3,4,6,7,11b-hexahydro-2H-pyrazino[2,1-a]isoquinoline-2-carbothioamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2022-02-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Development of Praziquantel sulphonamide derivatives as antischistosomal drugs.
J Enzyme Inhib Med Chem, 37, 2022
|
|
6DCE
 
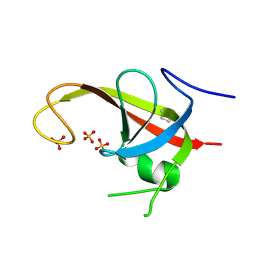 | | X-ray structure of FIP200 claw domain | | Descriptor: | RB1-inducible coiled-coil protein 1, SULFATE ION | | Authors: | Su, M.-Y, Hurley, J.H. | | Deposit date: | 2018-05-05 | | Release date: | 2019-03-06 | | Last modified: | 2019-05-01 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | FIP200 Claw Domain Binding to p62 Promotes Autophagosome Formation at Ubiquitin Condensates.
Mol. Cell, 74, 2019
|
|
4KTF
 
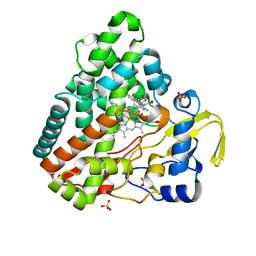 | | Crystal structure of Mycobacterium tuberculosis CYP121 in complex with 4,4'-(3-amino-1H-pyrazole-4,5-diyl)diphenol | | Descriptor: | 4,4'-(3-amino-1H-pyrazole-4,5-diyl)diphenol, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Hudson, S.A. | | Deposit date: | 2013-05-20 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Overcoming the Limitations of Fragment Merging: Rescuing a Strained Merged Fragment Series Targeting Mycobacterium tuberculosis CYP121.
Chemmedchem, 8, 2013
|
|
5HDI
 
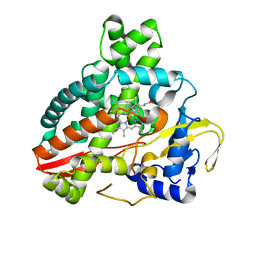 | | Structural characterization of CYP144A1, a Mycobacterium tuberculosis cytochrome P450 | | Descriptor: | Cytochrome P450 144, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chenge, J, Driscoll, M.D, McLean, K.J, Munro, A.W, Leys, D. | | Deposit date: | 2016-01-05 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural characterization of CYP144A1 - a cytochrome P450 enzyme expressed from alternative transcripts in Mycobacterium tuberculosis.
Sci Rep, 6, 2016
|
|
3HWO
 
 | |
6GMA
 
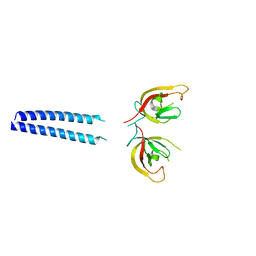 | |
6UH7
 
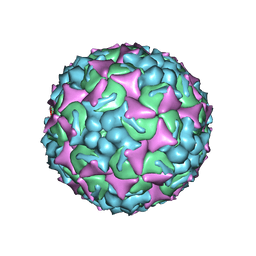 | | EV-A71 strain 11316 complexed with MADAL compound 30 | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Lee, H, Hafenstein, S. | | Deposit date: | 2019-09-26 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Scaffold Simplification Strategy Leads to a Novel Generation of Dual Human Immunodeficiency Virus and Enterovirus-A71 Entry Inhibitors.
J.Med.Chem., 63, 2020
|
|
1GU1
 
 | | Crystal structure of type II dehydroquinase from Streptomyces coelicolor complexed with 2,3-anhydro-quinic acid | | Descriptor: | 2,3 -ANHYDRO-QUINIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE, ... | | Authors: | Roszak, A.W, Robinson, D.A, Krell, T, Hunter, I.S, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-04-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure and Mechanism of the Type II Dehydroquinase from Streptomyces Coelicolor
Structure, 10, 2002
|
|
1GU0
 
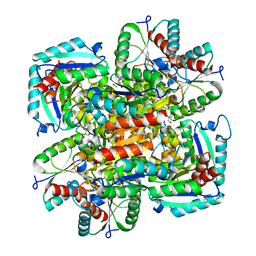 | | CRYSTAL STRUCTURE OF TYPE II DEHYDROQUINASE FROM STREPTOMYCES COELICOLOR | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Roszak, A.W, Krell, T, Robinson, D, Hunter, I.S, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-04-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure and Mechanism of the Type II Dehydroquinase from Streptomyces Coelicolor
Structure, 10, 2002
|
|
1GTZ
 
 | | Structure of STREPTOMYCES COELICOLOR TYPE II DEHYDROQUINASE R23A MUTANT IN COMPLEX WITH DEHYDROSHIKIMATE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE, 3-DEHYDROSHIKIMATE | | Authors: | Roszak, A.W, Krell, T, Robinson, D.A, Hunter, I.S, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure and Mechanism of the Type II Dehydroquinase from Streptomyces Coelicolor
Structure, 10, 2002
|
|
5C01
 
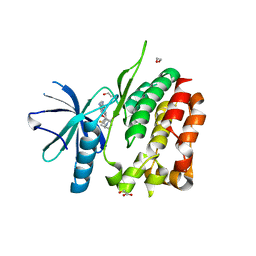 | | Crystal Structure of kinase | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, NON-RECEPTOR TYROSINE-PROTEIN KINASE TYK2, ... | | Authors: | Min, X, Wang, Z, Walker, N. | | Deposit date: | 2015-06-12 | | Release date: | 2015-09-16 | | Last modified: | 2015-11-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Characterization of the JH2 Pseudokinase Domain of JAK Family Tyrosine Kinase 2 (TYK2).
J.Biol.Chem., 290, 2015
|
|
5C03
 
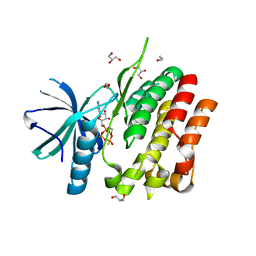 | | Crystal Structure of kinase | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Min, X, Wang, Z, Walker, N. | | Deposit date: | 2015-06-12 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Characterization of the JH2 Pseudokinase Domain of JAK Family Tyrosine Kinase 2 (TYK2).
J.Biol.Chem., 290, 2015
|
|
1V1J
 
 | | Crystal structure of type II Dehydroquintae Dehydratase from Streptomyces coelicolor in complex with 3-fluoro | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-ANHYDRO-3-FLUORO-QUINIC ACID, 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Roszak, A.W, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2004-04-16 | | Release date: | 2005-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | (1R,4S,5R)-3-Fluoro-1,4,5-Trihydroxy-2-Cyclohexene-1-Carboxylic Acid: The Fluoro Analogue of the Enolate Intermediate in the Reaction Catalyzed by Type II Dehydroquinases
Org.Biomol.Chem., 2, 2004
|
|
4MUK
 
 | |
7YJL
 
 | |
7YY0
 
 | |
7YY5
 
 | |
