6TQS
 
 | | The crystal structure of the MSP domain of human MOSPD2 in complex with the conventional FFAT motif of ORP1. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Di Mattia, T, Wendling, C, Cavarelli, J, Tomasetto, C, Alpy, F. | | Deposit date: | 2019-12-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts.
Embo J., 39, 2020
|
|
4UHP
 
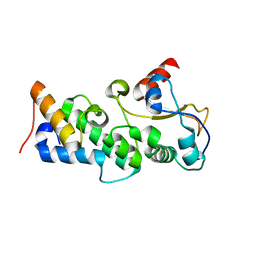 | | Crystal structure of the pyocin AP41 DNase-Immunity complex | | Descriptor: | BACTERIOCIN IMMUNITY PROTEIN, LARGE COMPONENT OF PYOCIN AP41 | | Authors: | Joshi, A, Chen, S, Wojdyla, J.A, Kaminska, R, Kleanthous, C. | | Deposit date: | 2015-03-25 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Ultra-High Affinity Protein-Protein Complexes of Pyocins S2 and Ap41 and Their Cognate Immunity Proteins from Pseudomonas Aeruginosa
J.Mol.Biol., 427, 2015
|
|
3SBY
 
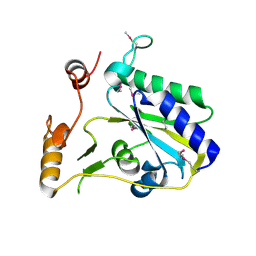 | | Crystal Structure of SeMet-Substituted Apo-MMACHC (1-244), a human B12 processing enzyme | | Descriptor: | Methylmalonic aciduria and homocystinuria type C protein | | Authors: | Koutmos, M, Gherasim, C, Smith, J.L, Banerjee, R. | | Deposit date: | 2011-06-06 | | Release date: | 2011-06-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural basis of multifunctionality in a vitamin B12-processing enzyme.
J.Biol.Chem., 286, 2011
|
|
4UHQ
 
 | | Crystal structure of the pyocin AP41 DNase | | Descriptor: | CITRIC ACID, LARGE COMPONENT OF PYOCIN AP41, NICKEL (II) ION | | Authors: | Joshi, A, Chen, S, Wojdyla, J.A, Kaminska, R, Kleanthous, C. | | Deposit date: | 2015-03-25 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of the Ultra-High Affinity Protein-Protein Complexes of Pyocins S2 and Ap41 and Their Cognate Immunity Proteins from Pseudomonas Aeruginosa
J.Mol.Biol., 427, 2015
|
|
6TJT
 
 | | Neuropilin2-b1 domain in a complex with the C-terminal VEGFC peptide | | Descriptor: | 1,2-ETHANEDIOL, Neuropilin-2, SULFATE ION, ... | | Authors: | Eldrid, C, Yu, L, Yelland, T, Fotinou, C, Djordjevic, S. | | Deposit date: | 2019-11-26 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | NRP2-b1 domain in a complex with the C-terminal VEGFC peptide
To Be Published
|
|
6TQU
 
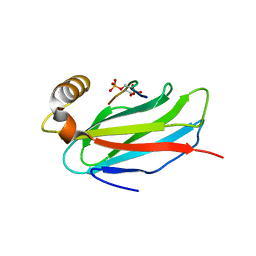 | | The crystal structure of the MSP domain of human MOSPD2 in complex with the Phospho-FFAT motif of STARD3. | | Descriptor: | Motile sperm domain-containing protein 2, SULFATE ION, StAR-related lipid transfer protein 3 | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Di Mattia, T, Wendling, C, Cavarelli, J, Tomasetto, C, Alpy, F. | | Deposit date: | 2019-12-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts.
Embo J., 39, 2020
|
|
7S02
 
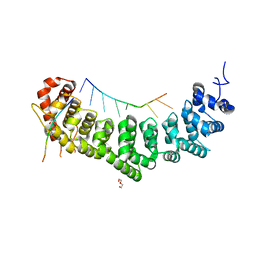 | |
4UDM
 
 | |
6TKK
 
 | | Neuropilin 1-b1 domain in a complex with the C-terminal VEGFB186 peptide | | Descriptor: | 1,2-ETHANEDIOL, ACE-ARG-PRO-GLN-PRO-ARG, Neuropilin-1 | | Authors: | Eldrid, C, Yu, L, Yelland, T, Fotinou, C, Djordjevic, S. | | Deposit date: | 2019-11-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Neuropilin 1-b1 domain in a complex with the C-terminal VEGFB186 peptide
To Be Published
|
|
6TQR
 
 | | The crystal structure of the MSP domain of human VAP-A in complex with the Phospho-FFAT motif of STARD3. | | Descriptor: | CHLORIDE ION, StAR-related lipid transfer protein 3, Vesicle-associated membrane protein-associated protein A | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Di Mattia, T, Wendling, C, Cavarelli, J, Tomasetto, C, Alpy, F. | | Deposit date: | 2019-12-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts.
Embo J., 39, 2020
|
|
6ZJD
 
 | | Crystal structure of human adenylate kinase 3, AK3, in complex with inhibitor ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GTP:AMP phosphotransferase AK3, ... | | Authors: | Grundstrom, C, Rogne, P, Wolf-Watz, M, Sauer-Eriksson, A.E. | | Deposit date: | 2020-06-28 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for GTP versus ATP Selectivity in the NMP Kinase AK3.
Biochemistry, 59, 2020
|
|
6TQT
 
 | | The crystal structure of the MSP domain of human MOSPD2. | | Descriptor: | 1,2-ETHANEDIOL, Motile sperm domain-containing protein 2, PHOSPHATE ION | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Di Mattia, T, Wendling, C, Cavarelli, J, Tomasetto, C, Alpy, F. | | Deposit date: | 2019-12-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts.
Embo J., 39, 2020
|
|
6ZJB
 
 | | Crystal structure of human adenylate kinase 3, AK3, in complex with inhibitor Gp5A | | Descriptor: | GTP:AMP phosphotransferase AK3, mitochondrial, MAGNESIUM ION, ... | | Authors: | Grundstrom, C, Rogne, P, Wolf-Watz, M, Sauer-Eriksson, A.E. | | Deposit date: | 2020-06-28 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.822 Å) | | Cite: | Structural Basis for GTP versus ATP Selectivity in the NMP Kinase AK3.
Biochemistry, 59, 2020
|
|
4WCW
 
 | | Ribosomal silencing factor during starvation or stationary phase (RsfS) from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, Ribosomal silencing factor RsfS | | Authors: | Li, X, Sun, Q, Jiang, C, Yang, K, Hung, L, Zhang, J, Sacchettini, J, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2014-09-05 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Ribosomal Silencing Factor Bound to Mycobacterium tuberculosis Ribosome.
Structure, 23, 2015
|
|
5JNB
 
 | | structure of GLD-2/RNP-8 complex | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Poly(A) RNA polymerase gld-2, ... | | Authors: | Nakel, K, Bonneau, F, Basquin, C, Eckmann, C.R, Conti, E. | | Deposit date: | 2016-04-29 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Structural basis for the antagonistic roles of RNP-8 and GLD-3 in GLD-2 poly(A)-polymerase activity.
Rna, 22, 2016
|
|
4V79
 
 | | E. coli 70S-fMetVal-tRNAVal-tRNAfMet complex in intermediate post-translocation state (post3b) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Blau, C, Bock, L.V, Schroder, G.F, Davydov, I, Fischer, N, Stark, H, Rodnina, M.V, Vaiana, A.C, Grubmuller, H. | | Deposit date: | 2013-10-14 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Energy barriers and driving forces in tRNA translocation through the ribosome.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4V82
 
 | | Crystal structure of cyanobacterial Photosystem II in complex with terbutryn | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Gabdulkhakov, A, Broser, M, Guskov, A, Kern, J, Glockner, C, Muh, F, Saenger, W, Zouni, A. | | Deposit date: | 2010-11-30 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of cyanobacterial photosystem II Inhibition by the herbicide terbutryn
J.Biol.Chem., 286, 2011
|
|
4UQT
 
 | | RRM-peptide structure in RES complex | | Descriptor: | PRE-MRNA-SPLICING FACTOR CWC26, U2 SNRNP COMPONENT IST3 | | Authors: | Tripsianes, K, Friberg, A, Barrandon, C, Seraphin, B, Sattler, M. | | Deposit date: | 2014-06-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A Novel Protein-Protein Interaction in the Res (Retention and Splicing) Complex.
J.Biol.Chem., 289, 2014
|
|
4UW5
 
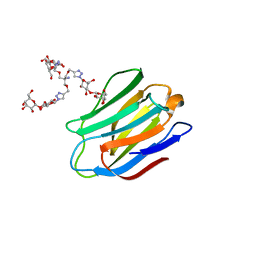 | | Human galectin-7 in complex with a galactose based dendron D2-2. | | Descriptor: | DENDRON D2-1, HUMAN GALECTIN-7 | | Authors: | Ramaswamy, S, Sleiman, M.H, Masuyer, G, Arbez-Gindre, C, Micha-Screttas, M, Calogeropoulou, T, Steele, B.R, Acharya, K.R. | | Deposit date: | 2014-08-08 | | Release date: | 2014-11-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis of Multivalent Galactose-Based Dendrimer Recognition by Human Galectin-7.
FEBS J., 282, 2015
|
|
4CML
 
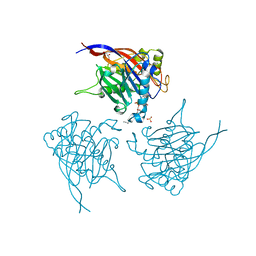 | | Crystal Structure of INPP5B in complex with Phosphatidylinositol 3,4- bisphosphate | | Descriptor: | 1,2-dioctanoyl phosphatidyl epi-inositol (3,4)-bisphosphate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tresaugues, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Ekblad, T, Flodin, S, Graslund, S, Karlberg, T, Moche, M, Nyman, T, Schuler, H, Silvander, C, Thorsell, A.G, Weigelt, J, Welin, M, Nordlund, P. | | Deposit date: | 2014-01-16 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Phosphoinositide Substrate Recognition, Catalysis, and Membrane Interactions in Human Inositol Polyphosphate 5-Phosphatases.
Structure, 22, 2014
|
|
6ERG
 
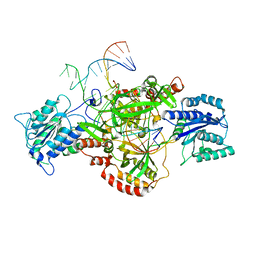 | | Complex of XLF and heterodimer Ku bound to DNA | | Descriptor: | DNA (21-MER), DNA (34-MER), Non-homologous end-joining factor 1, ... | | Authors: | Nemoz, C, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2017-10-18 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | XLF and APLF bind Ku80 at two remote sites to ensure DNA repair by non-homologous end joining.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4W66
 
 | |
4CMN
 
 | | Crystal structure of OCRL in complex with a phosphate ion | | Descriptor: | GLYCEROL, INOSITOL POLYPHOSPHATE 5-PHOSPHATASE OCRL-1, MAGNESIUM ION, ... | | Authors: | Tresaugues, L, Moche, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Ekblad, T, Flodin, S, Graslund, S, Karlberg, T, Nyman, T, Schuler, H, Silvander, C, Thorsell, A.G, Weigelt, J, Welin, M, Nordlund, P. | | Deposit date: | 2014-01-16 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Structural Basis for Phosphoinositide Substrate Recognition, Catalysis, and Membrane Interactions in Human Inositol Polyphosphate 5-Phosphatases.
Structure, 22, 2014
|
|
4UVA
 
 | | LSD1(KDM1A)-CoREST in complex with 1-Methyl-Tranylcypromine (1R,2S) | | Descriptor: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-2,3,4-trihydroxy-5-[(4aS)-4a-[(1S,3E)-3-imino-1-phenylbutyl]-7,8-dimethyl-2,4-dioxo-3,4,4a,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]pentyl dihydrogen diphosphate | | Authors: | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | Deposit date: | 2014-08-05 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
4W9F
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 5) | | Descriptor: | (4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-[4-(4-methyl-1,3-thiazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | van Molle, I, Hewitt, S, Galdeano, C, Gadd, M.S, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
