6ZJV
 
 | |
1Y5B
 
 | | Dianhydrosugar-based benzamidine, factor Xa specific inhibitor in complex with bovine trypsin mutant | | Descriptor: | 2,5-BIS-O-{4-[AMINO(IMINO)METHYL]PHENYL}-1,4:3,6-DIANHYDRO-D-GLUCITOL, CALCIUM ION, IMIDAZOLE, ... | | Authors: | Di Fenza, A, Heine, A, Klebe, G. | | Deposit date: | 2004-12-02 | | Release date: | 2005-12-13 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Understanding binding selectivity toward trypsin and factor Xa: the role of aromatic interactions
Chemmedchem, 2, 2007
|
|
6Z2O
 
 | | Crystal structure of wild type OgpA from Akkermansia muciniphila in P 21 21 21 | | Descriptor: | 1,2-ETHANEDIOL, O-glycan protease, ZINC ION | | Authors: | Trastoy, B, Naegali, A, Anso, I, Sjogren, J, Guerin, M.E. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Structural basis of mammalian mucin processing by the human gut O-glycopeptidase OgpA from Akkermansia muciniphila.
Nat Commun, 11, 2020
|
|
6Z2P
 
 | | Crystal structure of catalytic inactive OgpA from Akkermansia muciniphila in complex with an O-glycopeptide (glycodrosocin) substrate | | Descriptor: | CALCIUM ION, Glycodrosocin, O-glycan protease, ... | | Authors: | Trastoy, B, Naegali, A, Anso, I, Sjogren, J, Guerin, M.E. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural basis of mammalian mucin processing by the human gut O-glycopeptidase OgpA from Akkermansia muciniphila.
Nat Commun, 11, 2020
|
|
4M06
 
 | | Crystal Structure of Mutant Chlorite Dismutase from Candidatus Nitrospira defluvii W145F in Complex with Cyanide | | Descriptor: | 1,2-ETHANEDIOL, CYANIDE ION, Chlorite dismutase, ... | | Authors: | Hagmueller, A, Gysel, K, Djinovic-Carugo, K. | | Deposit date: | 2013-08-01 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Manipulating conserved heme cavity residues of chlorite dismutase: effect on structure, redox chemistry, and reactivity.
Biochemistry, 53, 2014
|
|
6ZJ3
 
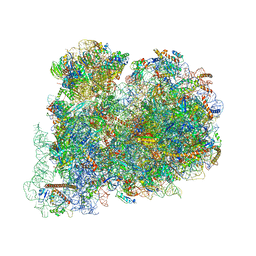 | | Cryo-EM structure of the highly atypical cytoplasmic ribosome of Euglena gracilis | | Descriptor: | 18S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Matzov, D, Halfon, H, Zimmerman, E, Rozenberg, H, Bashan, A, Gray, M.W, Yonath, A.E, Shalev-Benami, M. | | Deposit date: | 2020-06-27 | | Release date: | 2020-10-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Cryo-EM structure of the highly atypical cytoplasmic ribosome of Euglena gracilis.
Nucleic Acids Res., 48, 2020
|
|
2IJQ
 
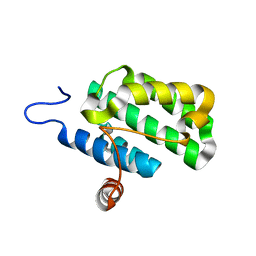 | | Crystal structure of protein rrnAC1037 from Haloarcula marismortui, Pfam DUF309 | | Descriptor: | Hypothetical protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Schwinn, K.D, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-30 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of the hypothetical Protein from Haloarcula marismortui
To be Published
|
|
6ZJP
 
 | |
1YI6
 
 | |
1YC1
 
 | | Crystal Structures of human HSP90alpha complexed with dihydroxyphenylpyrazoles | | Descriptor: | 4-(1,3-BENZODIOXOL-5-YL)-5-(5-ETHYL-2,4-DIHYDROXYPHENYL)-2H-PYRAZOLE-3-CARBOXYLIC ACID, Heat shock protein HSP 90-alpha | | Authors: | Kreusch, A, Han, S, Brinker, A, Zhou, V, Choi, H, He, Y, Lesley, S.A, Caldwell, J, Gu, X. | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of human HSP90alpha-complexed with dihydroxyphenylpyrazoles.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2IUF
 
 | | The structures of Penicillium vitale catalase: resting state, oxidised state (compound I) and complex with aminotriazole | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Murshudov, G, Borovik, A, Grebenko, A, Barynin, V, Vagin, A, Melik-Adamyan, W. | | Deposit date: | 2006-06-02 | | Release date: | 2006-07-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The Structures and Electronic Configuration of Compound I Intermediates of Helicobacter Pylori and Penicillium Vitale Catalases Determined by X-Ray Crystallography and Qm/Mm Density Functional Theory Calculations.
J.Am.Chem.Soc., 129, 2007
|
|
1Y59
 
 | | Dianhydrosugar-based benzamidine, factor Xa specific inhibitor in complex with bovine trypsin mutant | | Descriptor: | 2,5-BIS-O-{3-[AMINO(IMINO)METHYL]PHENYL}-1,4:3,6-DIANHYDRO-D-GLUCITOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Di Fenza, A, Heine, A, Klebe, G. | | Deposit date: | 2004-12-02 | | Release date: | 2005-12-13 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Understanding binding selectivity toward trypsin and factor Xa: the role of aromatic interactions
Chemmedchem, 2, 2007
|
|
1Y5U
 
 | | Dianhydrosugar-based benzamidine, factor Xa specific inhibitor in complex with bovine trypsin mutant | | Descriptor: | 2-O-{3-[AMINO(IMINO)METHYL]PHENYL}-5-O-{4-[AMINO(IMINO)METHYL]PHENYL}-1,4:3,6-DIANHYDRO-D-GLUCITOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Di Fenza, A, Heine, A, Klebe, G. | | Deposit date: | 2004-12-03 | | Release date: | 2005-12-13 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Understanding binding selectivity toward trypsin and factor Xa: the role of aromatic interactions
Chemmedchem, 2, 2007
|
|
2IHC
 
 | | Crystal structure of the bric-a-brac (BTB) domain of human BACH1 | | Descriptor: | Transcription regulator protein BACH1 | | Authors: | Amos, A.L, Turnbull, A.P, Bunkoczi, G, Papagrigoriou, E, Gorrec, F, Umeano, C, Bullock, A, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-09-26 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of the bric-a-brac (BTB) domain of human BACH1
To be Published
|
|
6TML
 
 | | Cryo-EM structure of Toxoplasma gondii mitochondrial ATP synthase hexamer, composite model | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit delta, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2019-12-04 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | ATP synthase hexamer assemblies shape cristae of Toxoplasma mitochondria.
Nat Commun, 12, 2021
|
|
2IDE
 
 | | Crystal Structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from Thermus Theromophilus HB8 | | Descriptor: | Molybdenum cofactor biosynthesis protein C, PHOSPHATE ION | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Baba, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-15 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from Thermus Theromophilus HB8
To be Published
|
|
4M07
 
 | | Crystal Structure of Mutant Chlorite Dismutase from Candidatus Nitrospira defluvii W145F | | Descriptor: | 1,2-ETHANEDIOL, Chlorite dismutase, GLYCEROL, ... | | Authors: | Hagmueller, A, Gysel, K, Djinovic-Carugo, K. | | Deposit date: | 2013-08-01 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Manipulating conserved heme cavity residues of chlorite dismutase: effect on structure, redox chemistry, and reactivity.
Biochemistry, 53, 2014
|
|
1YKA
 
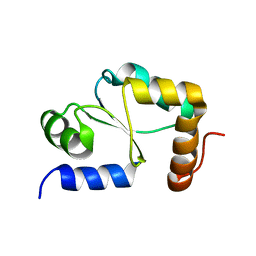 | | Solution structure of Grx4, a monothiol glutaredoxin from E. coli. | | Descriptor: | monothiol glutaredoxin ydhD | | Authors: | Fladvad, M, Bellanda, M, Fernandes, A.P, Andresen, C, Mammi, S, Holmgren, A, Vlamis-Gardikas, A, Sunnerhagen, M. | | Deposit date: | 2005-01-17 | | Release date: | 2005-04-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Molecular mapping of functionalities in the solution structure of reduced Grx4, a monothiol glutaredoxin from Escherichia coli.
J.Biol.Chem., 280, 2005
|
|
2III
 
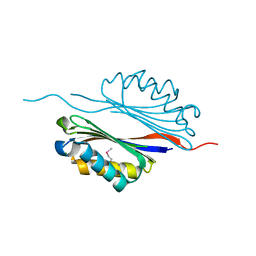 | | Crystal structure of the adenosylmethionine decarboxylase (aq_254) from aquifex aeolicus vf5 | | Descriptor: | CALCIUM ION, MAGNESIUM ION, S-adenosylmethionine decarboxylase proenzyme | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Baba, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-28 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the adenosylmethionine decarboxylase (aq_254) from aquifex aeolicus vf5
To be Published
|
|
1YJI
 
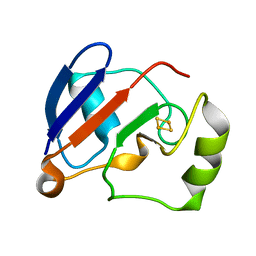 | | RDC-refined Solution NMR structure of reduced putidaredoxin | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Putidaredoxin | | Authors: | Jain, N.U, Tjioe, E, Savidor, A, Boulie, J. | | Deposit date: | 2005-01-14 | | Release date: | 2005-06-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Redox-dependent structural differences in putidaredoxin derived from homologous structure refinement via residual dipolar couplings.
Biochemistry, 44, 2005
|
|
1YNI
 
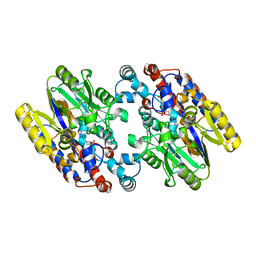 | | Crystal Structure of N-Succinylarginine Dihydrolase, AstB, bound to Substrate and Product, an Enzyme from the Arginine Catabolic Pathway of Escherichia coli | | Descriptor: | N~2~-(3-CARBOXYPROPANOYL)-L-ARGININE, POTASSIUM ION, Succinylarginine Dihydrolase | | Authors: | Tocilj, A, Schrag, J.D, Li, Y, Schneider, B.L, Reitzer, L, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of N-succinylarginine dihydrolase AstB, bound to substrate and product, an enzyme from the arginine catabolic pathway of Escherichia coli.
J.Biol.Chem., 280, 2005
|
|
2ISC
 
 | | Crystal structure of Purine Nucleoside Phosphorylase from Trichomonas vaginalis with DADMe-Imm-A | | Descriptor: | (3R,4R)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-(HYDROXYMETHYL)PYRROLIDIN-3-OL, PHOSPHATE ION, purine nucleoside phosphorylase | | Authors: | Rinaldo-Matthis, A, Almo, S.C, Schramm, V.L. | | Deposit date: | 2006-10-17 | | Release date: | 2007-06-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibition and structure of Trichomonas vaginalis purine nucleoside phosphorylase with picomolar transition state analogues
Biochemistry, 46, 2007
|
|
4D9W
 
 | |
4CRF
 
 | | Creating novel F1 inhibitors through fragment based lead generation and structure aided drug design | | Descriptor: | CHLORIDE ION, Coagulation factor XIa light chain, GLYCEROL, ... | | Authors: | Sandmark, J, Oster, L, Fjellstrom, O, Akkaya, S, Beisel, H.G, Eriksson, P.O, Erixon, K, Gustafsson, D, Jurva, U, Kang, D, Karis, D, Knecht, W, Nerme, V, Nilsson, I, Olsson, T, Redzic, A, Roth, R, Tigerstrom, A. | | Deposit date: | 2014-02-26 | | Release date: | 2015-02-11 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Creating Novel Activated Factor Xi Inhibitors Through Fragment Based Lead Generation and Structure Aided Drug Design.
Plos One, 10, 2015
|
|
4CIY
 
 | | Crystal structure of Mycobacterium tuberculosis type 2 dehydroquinase in complex with (1R,4R,5R)-1,4,5-trihydroxy-3-((1R)-1-hydroxy-2- phenyl)ethylcyclohex-2-en-1-carboxylic acid | | Descriptor: | (1R,4R,5R)-1,4,5-trihydroxy-3-[(1R)-1-hydroxy-2-phenyl]ethylcyclohex-2-ene-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE, CHLORIDE ION, ... | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Lamb, H, Hawkins, A.R, Blanco, B, Sedes, A, Peon, A, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2013-12-17 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exploring the water-binding pocket of the type II dehydroquinase enzyme in the structure-based design of inhibitors.
J. Med. Chem., 57, 2014
|
|
