2P69
 
 | | Crystal Structure of Human Pyridoxal Phosphate Phosphatase with PLP | | Descriptor: | CALCIUM ION, PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate phosphatase | | Authors: | Ramagopal, U.A, Freeman, J, Izuka, M, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
2P8E
 
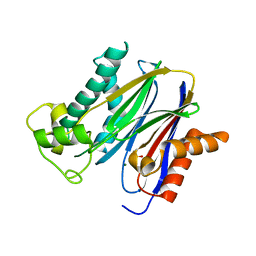 | | Crystal structure of the serine/threonine phosphatase domain of human PPM1B | | Descriptor: | MAGNESIUM ION, PPM1B beta isoform variant 6 | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Lau, C, Xu, W, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-22 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
1PVD
 
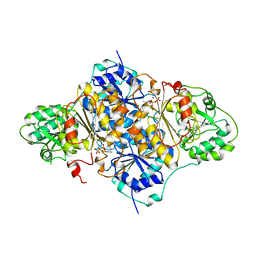 | |
1D0H
 
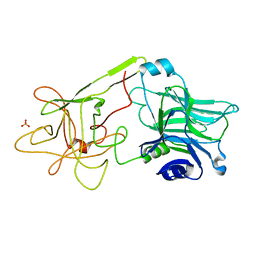 | | THE HC FRAGMENT OF TETANUS TOXIN COMPLEXED WITH N-ACETYL-GALACTOSAMINE | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, PROTEIN (TETANUS TOXIN HC), SULFATE ION | | Authors: | Emsley, P, Fotinou, C, Black, I, Fairweather, N.F, Charles, I.G, Watts, C, Hewitt, E, Isaacs, N.W. | | Deposit date: | 1999-09-10 | | Release date: | 2000-03-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structures of the H(C) fragment of tetanus toxin with carbohydrate subunit complexes provide insight into ganglioside binding.
J.Biol.Chem., 275, 2000
|
|
1PYD
 
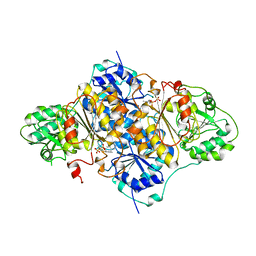 | |
8EUA
 
 | | Structure of SARS-CoV2 PLpro bound to a covalent inhibitor | | Descriptor: | Papain-like protease nsp3, SULFATE ION, ZINC ION, ... | | Authors: | Mathews, I.I, Pokhrel, S, Wakatsuki, S. | | Deposit date: | 2022-10-18 | | Release date: | 2023-04-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Potent and selective covalent inhibition of the papain-like protease from SARS-CoV-2.
Nat Commun, 14, 2023
|
|
7TEH
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-2 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-05 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
7TFR
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with NBH-2 | | Descriptor: | (1R,2S,5S)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-({1-[(2-methylpropane-2-sulfonyl)methyl]cyclohexyl}carbamoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-07 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
7TDU
 
 | | Joint X-ray/neutron structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-1 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxo(1-~2~H)pyrrolidin-3-yl]propan-2-yl}-3-{N-[tert-butyl(~2~H)carbamoyl]-3-methyl-L-(N-~2~H)valyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-(~2~H)carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-03 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
2OLA
 
 | | Crystal structure of O-succinylbenzoic acid synthetase from Staphylococcus aureus, cubic crystal form | | Descriptor: | O-succinylbenzoic acid synthetase | | Authors: | Patskovsky, Y, Sauder, J.M, Ozyurt, S, Wasserman, S.R, Smith, D, Dickey, M, Maletic, M, Reyes, C, Gheyi, T, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-18 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2OZT
 
 | | Crystal structure of O-succinylbenzoate synthase from Thermosynechococcus elongatus BP-1 | | Descriptor: | PHOSPHATE ION, SODIUM ION, Tlr1174 protein | | Authors: | Malashkevich, V.N, Bonanno, J, Toro, R, Sauder, J.M, Schwinn, K.D, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Gheyi, T, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-27 | | Release date: | 2007-03-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2NM1
 
 | | Structure of BoNT/B in complex with its protein receptor | | Descriptor: | Botulinum neurotoxin type B, Synaptotagmin-2 | | Authors: | Jin, R, Rummel, A, Binz, T, Brunger, A.T. | | Deposit date: | 2006-10-20 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Botulinum neurotoxin B recognizes its protein receptor with high affinity and specificity.
Nature, 444, 2006
|
|
3AFQ
 
 | | Crystal structure of the single-stranded DNA binding protein from Mycobacterium leprae (Form II) | | Descriptor: | Single-stranded DNA-binding protein | | Authors: | Kaushal, P.S, Singh, P, Sharma, A, Muniyappa, K, Vijayan, M. | | Deposit date: | 2010-03-10 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray and molecular-dynamics studies on Mycobacterium leprae single-stranded DNA-binding protein and comparison with other eubacterial SSB structures
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3AFP
 
 | | Crystal structure of the single-stranded DNA binding protein from Mycobacterium leprae (Form I) | | Descriptor: | CADMIUM ION, GLYCEROL, Single-stranded DNA-binding protein | | Authors: | Kaushal, P.S, Singh, P, Sharma, A, Muniyappa, K, Vijayan, M. | | Deposit date: | 2010-03-10 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray and molecular-dynamics studies on Mycobacterium leprae single-stranded DNA-binding protein and comparison with other eubacterial SSB structures
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3A5U
 
 | | Promiscuity and specificity in DNA binding to SSB: Insights from the structure of the Mycobacterium smegmatis SSB-ssDNA complex | | Descriptor: | DNA (31-MER), Single-stranded DNA-binding protein | | Authors: | Kaushal, P.S, Manjunath, G.P, Sekar, K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2009-08-12 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Promiscuity and specificity in DNA binding to SSB: Insights from the structure of the Mycobacterium smegmatis SSB-ssDNA complex.
To be Published, 2009
|
|
6V2F
 
 | | Crystal structure of the HIV capsid hexamer bound to the small molecule long-acting inhibitor, GS-6207 | | Descriptor: | HIV-1 capsid, N-[(1S)-1-(3-{4-chloro-3-[(methylsulfonyl)amino]-1-(2,2,2-trifluoroethyl)-1H-indazol-7-yl}-6-[3-methyl-3-(methylsulfonyl)but-1-yn-1-yl]pyridin-2-yl)-2-(3,5-difluorophenyl)ethyl]-2-[(3bS,4aR)-5,5-difluoro-3-(trifluoromethyl)-3b,4,4a,5-tetrahydro-1H-cyclopropa[3,4]cyclopenta[1,2-c]pyrazol-1-yl]acetamide | | Authors: | Appleby, T.C, Link, J.O, Yant, S.R, Villasenor, A.G, Somoza, J.R, Hu, E.Y, Schroeder, S.D, Cihlar, T. | | Deposit date: | 2019-11-22 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Clinical targeting of HIV capsid protein with a long-acting small molecule.
Nature, 584, 2020
|
|
4P0T
 
 | |
4R7M
 
 | |
4R5V
 
 | |
4R5X
 
 | |
4R76
 
 | |
4R5T
 
 | |
4R6T
 
 | |
1BJR
 
 | | COMPLEX FORMED BETWEEN PROTEOLYTICALLY GENERATED LACTOFERRIN FRAGMENT AND PROTEINASE K | | Descriptor: | CALCIUM ION, LACTOFERRIN, PROTEINASE K | | Authors: | Singh, T.P, Sharma, S, Karthikeyan, S, Betzel, C, Bhatia, K.L. | | Deposit date: | 1998-06-27 | | Release date: | 1998-11-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of a complex formed between proteolytically-generated lactoferrin fragment and proteinase K.
Proteins, 33, 1998
|
|
4OUC
 
 | | Structure of human haspin in complex with histone H3 substrate | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 1,2-ETHANEDIOL, Histone H3.2, ... | | Authors: | Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-02-15 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modulation of the chromatin phosphoproteome by the haspin protein kinase.
Mol Cell Proteomics, 13, 2014
|
|
