7VVN
 
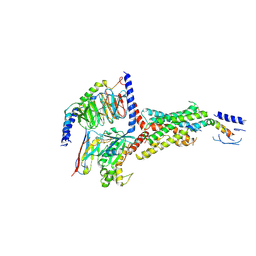 | | PTH-bound human PTH1R in complex with Gs (class4) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Kobayashi, K, Kusakizako, T, Miyauchi, H, Tomita, A, Kobayashi, K, Shihoya, W, Yamashita, K, Nishizawa, T, Kato, H.E, Nureki, O. | | Deposit date: | 2021-11-06 | | Release date: | 2022-08-03 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Endogenous ligand recognition and structural transition of a human PTH receptor.
Mol.Cell, 82, 2022
|
|
7BM6
 
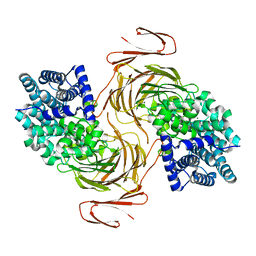 | | Structure-function analysis of a new PL17 oligoalginate lyase from the marine bacterium Zobellia galactanivorans DsijT | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid, Alginate lyase, family PL17, ... | | Authors: | Czjzek, M, Roret, T, Jouanneau, D, Le Duff, N, Jeudy, A. | | Deposit date: | 2021-01-19 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure-function analysis of a new PL17 oligoalginate lyase from the marine bacterium Zobellia galactanivorans DsijT.
Glycobiology, 31, 2021
|
|
7VVK
 
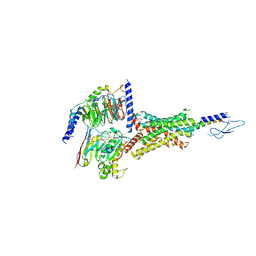 | | PTH-bound human PTH1R in complex with Gs (class1) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Kobayashi, K, Kusakizako, T, Miyauchi, H, Tomita, A, Kobayashi, K, Shihoya, W, Yamashita, K, Nishizawa, T, Kato, H.E, Nureki, O. | | Deposit date: | 2021-11-06 | | Release date: | 2022-08-03 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Endogenous ligand recognition and structural transition of a human PTH receptor.
Mol.Cell, 82, 2022
|
|
7VVM
 
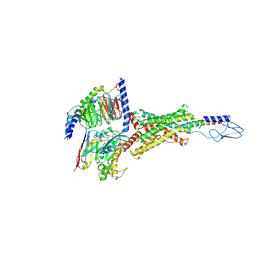 | | PTH-bound human PTH1R in complex with Gs (class3) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Kobayashi, K, Kusakizako, T, Miyauchi, H, Tomita, A, Kobayashi, K, Shihoya, W, Yamashita, K, Nishizawa, T, Kato, H.E, Nureki, O. | | Deposit date: | 2021-11-06 | | Release date: | 2022-08-03 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Endogenous ligand recognition and structural transition of a human PTH receptor.
Mol.Cell, 82, 2022
|
|
5AE3
 
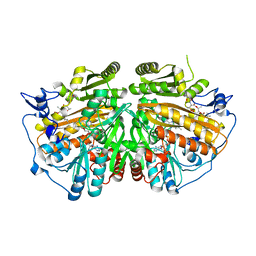 | | Ether Lipid-Generating Enzyme AGPS in complex with antimycin A | | Descriptor: | ALKYLDIHYDROXYACETONEPHOSPHATE SYNTHASE, PEROXISOMAL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Piano, V, Benjamin, D.I, Valente, S, Nenci, S, Marrocco, B, Mai, A, Aliverti, A, Nomura, D.K, Mattevi, A. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of Inhibitors for the Ether Lipid-Generating Enzyme Agps as Anti-Cancer Agents.
Acs Chem.Biol., 10, 2015
|
|
7VVJ
 
 | | PTHrP-bound human PTH1R in complex with Gs | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Kobayashi, K, Kusakizako, T, Miyauchi, H, Tomita, A, Kobayashi, K, Shihoya, W, Yamashita, K, Nishizawa, T, Kato, H.E, Nureki, O. | | Deposit date: | 2021-11-06 | | Release date: | 2022-08-03 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Endogenous ligand recognition and structural transition of a human PTH receptor.
Mol.Cell, 82, 2022
|
|
7VVO
 
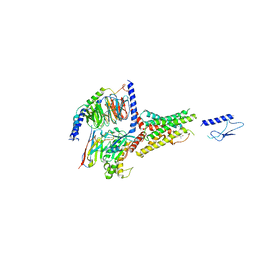 | | PTH-bound human PTH1R in complex with Gs (class5) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Kobayashi, K, Kusakizako, T, Miyauchi, H, Tomita, A, Kobayashi, K, Shihoya, W, Yamashita, K, Nishizawa, T, Kato, H.E, Nureki, O. | | Deposit date: | 2021-11-06 | | Release date: | 2022-08-03 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Endogenous ligand recognition and structural transition of a human PTH receptor.
Mol.Cell, 82, 2022
|
|
7BJY
 
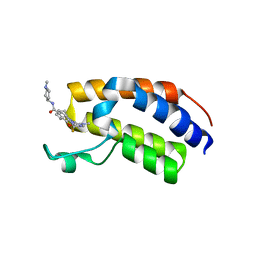 | | Crystal structure of the first bromodomain (BD1) of human BRDT bound to Ro3280 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(9-cyclopentyl-7,7-difluoro-5-methyl-6-oxo-6,7,8,9-tetrahydro-5H-pyrimido[4,5-b][1,4]diazepin-2-yl)amino]-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Isoform 2 of Bromodomain testis-specific protein | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2021-01-14 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
4KAH
 
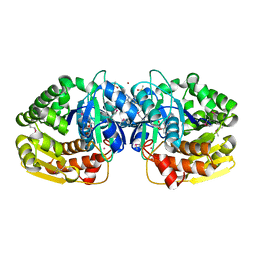 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with 4-bromo-1H-pyrazole | | Descriptor: | 4-bromo-1H-pyrazole, ADENOSINE, BROMIDE ION, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-22 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with 4-bromo-1H-pyrazole
To be Published
|
|
4GWB
 
 | | Crystal structure of putative Peptide methionine sulfoxide reductase from Sinorhizobium meliloti 1021 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Peptide methionine sulfoxide reductase MsrA 3 | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-09-01 | | Release date: | 2012-09-19 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of putative Peptide methionine sulfoxide reductase from Sinorhizobium meliloti 1021
To be Published
|
|
2X57
 
 | | Crystal structure of the extracellular domain of human Vasoactive intestinal polypeptide receptor 2 | | Descriptor: | VASOACTIVE INTESTINAL POLYPEPTIDE RECEPTOR 2 | | Authors: | Pike, A.C.W, Barr, A.J, Quigley, A, Burgess Brown, N, de Riso, A, Bullock, A, Berridge, G, Muniz, J.R.C, Chaikaud, A, Vollmar, M, Krojer, T, Ugochukwu, E, von Delft, F, Edwards, A, Arrowsmith, C.H, Weigelt, J, Bountra, C, Carpenter, E.P. | | Deposit date: | 2010-02-05 | | Release date: | 2010-03-09 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Extracellular Domain of Human Vasoactive Intestinal Polypeptide Receptor 2
To be Published
|
|
7ZRY
 
 | |
4HAD
 
 | | Crystal structure of probable oxidoreductase protein from Rhizobium etli CFN 42 | | Descriptor: | Probable oxidoreductase protein, SODIUM ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-09-26 | | Release date: | 2012-10-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of probable oxidoreductase protein from Rhizobium etli CFN 42
To be Published
|
|
5AQ9
 
 | | DARPin-based Crystallization Chaperones exploit Molecular Geometry as a Screening Dimension in Protein Crystallography | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, OFF7_DB08V4 | | Authors: | Batyuk, A, Wu, Y, Honegger, A, Heberling, M, Plueckthun, A. | | Deposit date: | 2015-09-21 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Darpin-Based Crystallization Chaperones Exploit Molecular Geometry as a Screening Dimension in Protein Crystallography
J.Mol.Biol., 428, 2016
|
|
4QBK
 
 | | Crystal structure of the complex of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa with amino acyl-tRNA analogue at 1.77 Angstrom resolution | | Descriptor: | 3'-deoxy-3'-[(O-methyl-L-tyrosyl)amino]adenosine, GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Singh, A, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-05-08 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and binding studies of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa provide a platform for the structure-based inhibitor design against peptidyl-tRNA hydrolase
Biochem.J., 463, 2014
|
|
5E2M
 
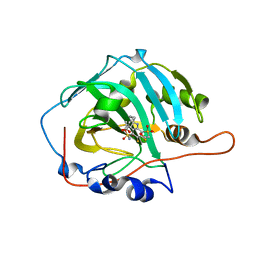 | | Crystal structure of human carbonic anhydrase isozyme I with 3-(cyclooctylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 3-(cyclooctylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, ACETATE ION, Carbonic anhydrase 1, ... | | Authors: | Manakova, E, Smirnov, A, Grazulis, S. | | Deposit date: | 2015-10-01 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Intrinsic Thermodynamics and Structures of 2,4- and 3,4-Substituted Fluorinated Benzenesulfonamides Binding to Carbonic Anhydrases.
ChemMedChem, 12, 2017
|
|
5EBK
 
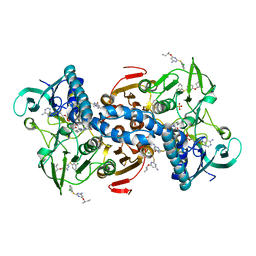 | |
5MA6
 
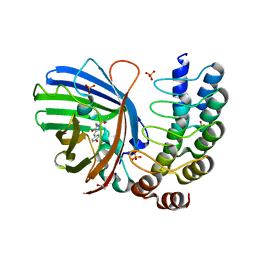 | | GFP-binding DARPin 3G124nc | | Descriptor: | 1,2-ETHANEDIOL, 3G124nc, Green fluorescent protein, ... | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
5MAK
 
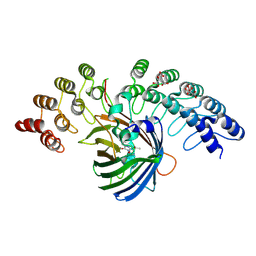 | | GFP-binding DARPin fusion gc_R7 | | Descriptor: | CITRIC ACID, Green fluorescent protein, R7 | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
4I5F
 
 | | Crystal structure of Ralstonia sp. alcohol dehydrogenase mutant N15G, G37D, R38V, R39S | | Descriptor: | Alclohol dehydrogenase/short-chain dehydrogenase | | Authors: | Jarasch, A, Lerchner, A, Meining, W, Schiefner, A, Skerra, A. | | Deposit date: | 2012-11-28 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic analysis and structure-guided engineering of NADPH-dependent Ralstonia sp. Alcohol dehydrogenase toward NADH cosubstrate specificity.
Biotechnol.Bioeng., 110, 2013
|
|
5MNA
 
 | | Cationic trypsin in complex with aniline (deuterated sample at 295 K) | | Descriptor: | ANILINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2016-12-13 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.441 Å) | | Cite: | Charges Shift Protonation: Neutron Diffraction Reveals that Aniline and 2-Aminopyridine Become Protonated Upon Binding to Trypsin.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
8UCZ
 
 | |
8PTC
 
 | | COMPLEX CRYSTAL STRUCTURE OF MUTANT HUMAN MONOGLYCERIDE LIPASE WITH COMPOUND 5d | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3~{R},4~{S})-2-oxidanylidene-3,4-diphenyl-azetidin-1-yl]piperidine-1-carbaldehyde, Monoglyceride lipase | | Authors: | Butini, S, Benz, J, Grether, U, Leibrock, L, Papa, A, Maramai, S, Carullo, G, Federico, S, Grillo, A, Di Guglielmo, B, Lamponi, S, Gemma, S, Campiani, G. | | Deposit date: | 2023-07-14 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Development of Potent and Selective Monoacylglycerol Lipase Inhibitors. SARs, Structural Analysis, and Biological Characterization.
J.Med.Chem., 67, 2024
|
|
8PTR
 
 | | COMPLEX CRYSTAL STRUCTURE OF MUTANT HUMAN MONOGLYCERIDE LIPASE WITH COMPOUND 5r | | Descriptor: | (3~{R},4~{S})-4-(1,3-benzodioxol-5-yl)-1-[1-(benzotriazol-1-ylcarbonyl)piperidin-4-yl]-3-(3-fluorophenyl)azetidin-2-one, 1,2-ETHANEDIOL, Monoglyceride lipase | | Authors: | Butini, S, Benz, J, Grether, U, Leibrock, L, Papa, A, Maramai, S, Carullo, G, Federico, S, Grillo, A, Di Guglielmo, B, Lamponi, S, Gemma, S, Campiani, G. | | Deposit date: | 2023-07-14 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Development of Potent and Selective Monoacylglycerol Lipase Inhibitors. SARs, Structural Analysis, and Biological Characterization.
J.Med.Chem., 67, 2024
|
|
7BBR
 
 | | Crystal structure of the sugar acid binding protein DctPAm from Advenella mimigardefordensis strain DPN7T | | Descriptor: | 2-KETO-3-DEOXYGLUCONATE, Putative TRAP transporter solute receptor DctP | | Authors: | Schaefer, L, Meinert, C, Kobus, S, Hoeppner, A, Smits, S.H, Steinbuechel, A. | | Deposit date: | 2020-12-18 | | Release date: | 2021-03-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the sugar acid-binding protein CxaP from a TRAP transporter in Advenella mimigardefordensis strain DPN7 T .
Febs J., 288, 2021
|
|
