6C6Z
 
 | |
7WTJ
 
 | | SARS-CoV-2 Omicron variant spike RBD in complex with Fab XGv286 | | Descriptor: | Heavy chain of XGv286, Light chain of XGv286, Spike protein S1 | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-02-04 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTK
 
 | | SARS-CoV-2 Omicron variant spike in complex with Fab XGv286 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of XGv286, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-02-04 | | Release date: | 2022-12-14 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTF
 
 | | SARS-CoV-2 Omicron variant spike in complex with Fab XGv051 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of XGv051, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-02-04 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTG
 
 | | SARS-CoV-2 Omicron variant spike RBD in complex with Fab XGv051 | | Descriptor: | Heavy chain of XGv051, Light chain of XGv051, Spike protein S1 | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-02-04 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTH
 
 | | SARS-CoV-2 Omicron variant spike RBD in complex with Fab XGv264 | | Descriptor: | Heavy chain of XGv264, Light chain of XGv264, Spike protein S1 | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-02-04 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTI
 
 | | SARS-CoV-2 Omicron variant spike in complex with Fab XGv264 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of XGv264, Light chain of XGv264, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-02-04 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7LRT
 
 | |
7LRS
 
 | |
6C6Y
 
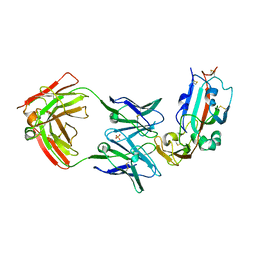 | | Crystal structure of Middle-East Respiratory Syndrome (MERS) coronavirus neutralizing antibody JC57-14 isolated from a vaccinated rhesus macaque in complex with MERS Receptor Binding Domain | | Descriptor: | JC57-14 Heavy chain, JC57-14 Light chain, SULFATE ION, ... | | Authors: | Joyce, M.G, Mascola, J.R, Graham, B.S, Kwong, P.D. | | Deposit date: | 2018-01-19 | | Release date: | 2018-03-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Importance of Neutralizing Monoclonal Antibodies Targeting Multiple Antigenic Sites on the Middle East Respiratory Syndrome Coronavirus Spike Glycoprotein To Avoid Neutralization Escape.
J. Virol., 92, 2018
|
|
6C6X
 
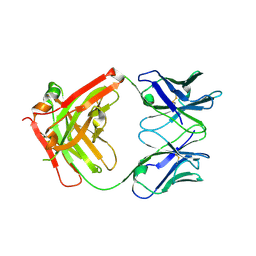 | | Crystal structure of Middle-East Respiratory Syndrome (MERS) coronavirus neutralizing antibody JC57-14 isolated from a vaccinated rhesus macaque. | | Descriptor: | JC57-14 Heavy chain, JC57-14 Light chain | | Authors: | Joyce, M.G, Mascola, J.R, Graham, B.S, Kwong, P.D. | | Deposit date: | 2018-01-19 | | Release date: | 2018-02-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Importance of Neutralizing Monoclonal Antibodies Targeting Multiple Antigenic Sites on the Middle East Respiratory Syndrome Coronavirus Spike Glycoprotein To Avoid Neutralization Escape.
J. Virol., 92, 2018
|
|
7MLZ
 
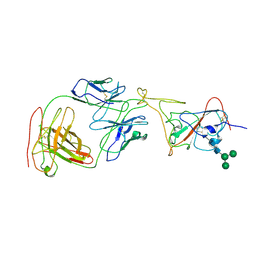 | | Cryo-EM structure of SARS-CoV-2 spike in complex with neutralizing antibody B1-182.1 that targets the receptor-binding domain | | Descriptor: | B1-182.1 Fab heavy chain, B1-182.1 Fab light chain, Spike protein S1, ... | | Authors: | Zhou, T, Tsybovsky, T, Kwong, P.D. | | Deposit date: | 2021-04-29 | | Release date: | 2021-07-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants.
Science, 373, 2021
|
|
7MM0
 
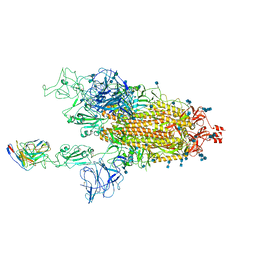 | | Cryo-EM structure of SARS-CoV-2 spike in complex with neutralizing antibody B1-182.1 that targets the receptor-binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, B1-182.1 Fab heavy chain, ... | | Authors: | Zhou, T, Tsybovsky, T, Kwong, P.D. | | Deposit date: | 2021-04-29 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants.
Science, 373, 2021
|
|
4ZZ3
 
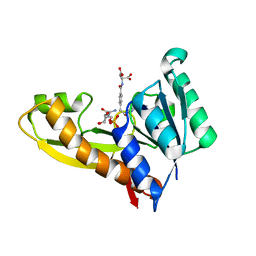 | | Human GAR transformylase in complex with GAR and pemetrexed | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-{4-[2-(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)ethyl]benzoyl}-L-glutamic acid, Trifunctional purine biosynthetic protein adenosine-3 | | Authors: | Deis, S.M, Dann III, C.E. | | Deposit date: | 2015-05-22 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | 6-Substituted Pyrrolo[2,3-d]pyrimidine Thienoyl Regioisomers as Targeted Antifolates for Folate Receptor alpha and the Proton-Coupled Folate Transporter in Human Tumors.
J.Med.Chem., 58, 2015
|
|
4ZZ1
 
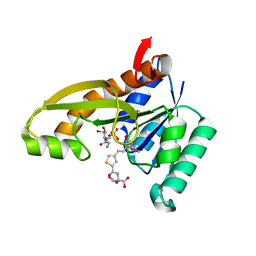 | | HUMAN GAR TRANSFORMYLASE IN COMPLEX WITH GAR AND (S)-2-({4-[3-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-6-YL)-PROPYL]-THIOPHENE-2-CARBONYL}-AMINO)-PENTANEDIOIC ACID | | Descriptor: | (S)-2-({4-[3-(2-Amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-6-yl)-propyl]-thiophene-2-carbonyl}-amino)-pentanedioic acid, GLYCINAMIDE RIBONUCLEOTIDE, Trifunctional purine biosynthetic protein adenosine-3 | | Authors: | Deis, S.M, Dann III, C.E. | | Deposit date: | 2015-05-22 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | 6-Substituted Pyrrolo[2,3-d]pyrimidine Thienoyl Regioisomers as Targeted Antifolates for Folate Receptor alpha and the Proton-Coupled Folate Transporter in Human Tumors.
J.Med.Chem., 58, 2015
|
|
4ZZ2
 
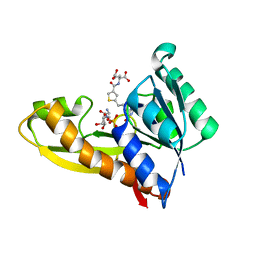 | | HUMAN GAR TRANSFORMYLASE IN COMPLEX WITH GAR AND (S)-2-({5-[3-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-6-YL)-PROPYL]-THIOPHENE-3-CARBONYL}-AMINO)-PENTANEDIOIC ACID | | Descriptor: | (S)-2-({5-[3-(2-Amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-6-yl)-propyl]-thiophene-3-carbonyl}-amino)-pentanedioic acid, GLYCINAMIDE RIBONUCLEOTIDE, Trifunctional purine biosynthetic protein adenosine-3 | | Authors: | Deis, S.M, Dann III, C.E. | | Deposit date: | 2015-05-22 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | 6-Substituted Pyrrolo[2,3-d]pyrimidine Thienoyl Regioisomers as Targeted Antifolates for Folate Receptor alpha and the Proton-Coupled Folate Transporter in Human Tumors.
J.Med.Chem., 58, 2015
|
|
7DWC
 
 | | Bacteroides thetaiotaomicron VPI5482 BTAxe1 | | Descriptor: | Xylanase | | Authors: | Wang, L.Y, Wang, Y.L, Xin, F.J, Sun, L.C. | | Deposit date: | 2021-01-17 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Rational Design for Broadened Substrate Specificity and Enhanced Activity of a Novel Acetyl Xylan Esterase from Bacteroides thetaiotaomicron.
J.Agric.Food Chem., 69, 2021
|
|
7TYJ
 
 | | Cryo-EM Structure of insulin receptor-related receptor (IRR) in apo-state captured at pH 7. The 3D refinement was focused on one of two halves with C1 symmetry applied | | Descriptor: | Insulin receptor-related protein | | Authors: | Wang, L.W, Hall, C, Li, J, Choi, E, Bai, X.C. | | Deposit date: | 2022-02-13 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of the alkaline pH-dependent activation of insulin receptor-related receptor.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TYK
 
 | | Cryo-EM Structure of insulin receptor-related receptor (IRR) in apo-state captured at pH 7. The 3D refinement was applied with C2 symmetry | | Descriptor: | Insulin receptor-related protein | | Authors: | Wang, L.W, Hall, C, Li, J, Choi, E, Bai, X.C. | | Deposit date: | 2022-02-13 | | Release date: | 2023-02-15 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of the alkaline pH-dependent activation of insulin receptor-related receptor.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TYM
 
 | | Cryo-EM Structure of insulin receptor-related receptor (IRR) in active-state captured at pH 9. The 3D refinement was applied with C2 symmetry | | Descriptor: | Insulin receptor-related protein | | Authors: | Wang, L.W, Hall, C, Li, J, Choi, E, Bai, X.C. | | Deposit date: | 2022-02-13 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of the alkaline pH-dependent activation of insulin receptor-related receptor.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6EGE
 
 | |
6EGF
 
 | |
6EGD
 
 | |
1BM5
 
 | |
4J6O
 
 | | Crystal Structure of the Phosphatase Domain of C. thermocellum (Bacterial) PnkP | | Descriptor: | CITRIC ACID, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Wang, L, Smith, P, Shuman, S. | | Deposit date: | 2013-02-11 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of the 2',3' phosphatase component of the bacterial Pnkp-Hen1 RNA repair system.
Nucleic Acids Res., 41, 2013
|
|
