4KBN
 
 | | human dihydrofolate reductase complexed with NADPH and 5-{3-[3-(3,5-pyrimidine)]-phenyl-prop-1-yn-1-yl}-6-ethyl-pyrimidine-2,4diamine | | Descriptor: | 6-ethyl-5-{3-[3-(pyrimidin-5-yl)phenyl]prop-1-yn-1-yl}pyrimidine-2,4-diamine, AMMONIUM ION, CHLORIDE ION, ... | | Authors: | Lamb, K.M, Anderson, A.C. | | Deposit date: | 2013-04-23 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Elucidating features that drive the design of selective antifolates using crystal structures of human dihydrofolate reductase.
Biochemistry, 52, 2013
|
|
4KFJ
 
 | | Human dihydrofolate reductase complexed with NADPH and 5-{3-[3-methoxy-5-(isoquin-5-yl)phenyl]prop-1-yn-1-yl}6-ethylprimidine-2,4-diamine | | Descriptor: | 6-ethyl-5-{3-[3-(isoquinolin-5-yl)-5-methoxyphenyl]prop-1-yn-1-yl}pyrimidine-2,4-diamine, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Lamb, K.M, Anderson, A.C. | | Deposit date: | 2013-04-26 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Elucidating features that drive the design of selective antifolates using crystal structures of human dihydrofolate reductase.
Biochemistry, 52, 2013
|
|
4KD7
 
 | | Human dihydrofolate reductase complexed with NADPH and 5-{3-[3-methoxy-5(pyridine-4-yl)phenyl]prop-1-yn-1-yl}-6-ethyl-pyrimidine-2,4-diamine | | Descriptor: | 6-ethyl-5-{3-[3-methoxy-5-(pyridin-4-yl)phenyl]prop-1-yn-1-yl}pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lamb, K.M, Anderson, A.C. | | Deposit date: | 2013-04-24 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.715 Å) | | Cite: | Elucidating features that drive the design of selective antifolates using crystal structures of human dihydrofolate reductase.
Biochemistry, 52, 2013
|
|
8J6G
 
 | |
8EIH
 
 | | Cryo-EM structure of human DNMT3B homo-tetramer (form I) | | Descriptor: | DNA (cytosine-5)-methyltransferase 3B, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Lu, J.W, Song, J.K. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural basis for the allosteric regulation and dynamic assembly of DNMT3B.
Nucleic Acids Res., 51, 2023
|
|
8EIK
 
 | | Cryo-EM structure of human DNMT3B homo-hexamer | | Descriptor: | DNA (cytosine-5)-methyltransferase 3B, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Lu, J.W, Song, J.K. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis for the allosteric regulation and dynamic assembly of DNMT3B.
Nucleic Acids Res., 51, 2023
|
|
8EIJ
 
 | | Cryo-EM structure of human DNMT3B homo-trimer | | Descriptor: | DNA (cytosine-5)-methyltransferase 3B, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Lu, J.W, Song, J.K. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural basis for the allosteric regulation and dynamic assembly of DNMT3B.
Nucleic Acids Res., 51, 2023
|
|
8EII
 
 | | Cryo-EM structure of human DNMT3B homo-tetramer (form II) | | Descriptor: | DNA (cytosine-5)-methyltransferase 3B, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Lu, J.W, Song, J.K. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural basis for the allosteric regulation and dynamic assembly of DNMT3B.
Nucleic Acids Res., 51, 2023
|
|
8F0V
 
 | | Lipocalin-like Milk protein-2 - E38A mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Milk protein, ZINC ION | | Authors: | Subramanian, R, KanagaVijayan, D. | | Deposit date: | 2022-11-04 | | Release date: | 2023-08-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Variability in phenylalanine side chain conformations facilitates broad substrate tolerance of fatty acid binding in cockroach milk proteins.
Plos One, 18, 2023
|
|
8F0Y
 
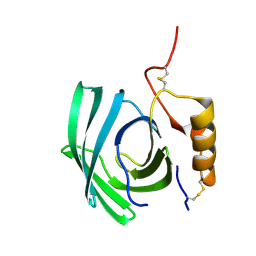 | | Lipocalin-like Milk protein-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Milk protein | | Authors: | Subramanian, R, KanagaVijayan, D, Shantakumar, R.P.S. | | Deposit date: | 2022-11-04 | | Release date: | 2023-08-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Variability in phenylalanine side chain conformations facilitates broad substrate tolerance of fatty acid binding in cockroach milk proteins.
Plos One, 18, 2023
|
|
6OMF
 
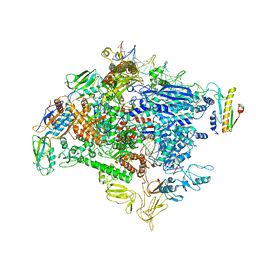 | | CryoEM structure of SigmaS-transcription initiation complex with activator Crl | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Jaramillo Cartagena, A, Darst, S.A, Campbell, E.A. | | Deposit date: | 2019-04-18 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis for transcription activation by Crl through tethering of sigmaSand RNA polymerase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2LU1
 
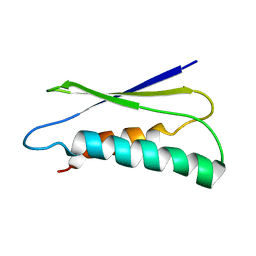 | | pfsub2 solution NMR structure | | Descriptor: | Subtilase | | Authors: | He, Y, Chen, Y, Ruan, B, O'Brochta, D, Bryan, P, Orban, J. | | Deposit date: | 2012-06-06 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a sheddase inhibitor prodomain from the malarial parasite Plasmodium falciparum.
Proteins, 80, 2012
|
|
7RRW
 
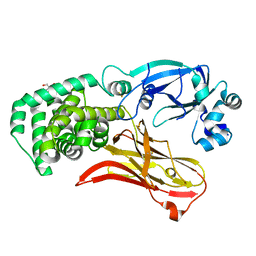 | | Monomeric CRM197 expressed in E. coli | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Diphtheria toxin | | Authors: | Gallagher, D.T, Lees, A. | | Deposit date: | 2021-08-10 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Monomeric crystal structure of the vaccine carrier protein CRM 197 and implications for vaccine development.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
9GK2
 
 | |
9KKH
 
 | | High resolution structure of Ferredoxin-NADP+ reductase from maize root - Reduced form, low X-ray dose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, ... | | Authors: | Uenaka, M, Ohnishi, Y, Tanaka, H, Kurisu, G. | | Deposit date: | 2024-11-13 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Redox-dependent hydrogen-bond network rearrangement of ferredoxin-NADP + reductase revealed by high-resolution X-ray and neutron crystallography.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
9KK7
 
 | | Neutron structure of Ferredoxin-NADP+ reductase from maize root -Reduced form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, ... | | Authors: | Uenaka, M, Ohnishi, Y, Tanaka, H, Kurisu, G. | | Deposit date: | 2024-11-13 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-12 | | Method: | NEUTRON DIFFRACTION (1.8 Å) | | Cite: | Redox-dependent hydrogen-bond network rearrangement of ferredoxin-NADP + reductase revealed by high-resolution X-ray and neutron crystallography.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
9KKC
 
 | | Neutron structure of Ferredoxin-NADP+ reductase from maize root -Oxidized form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, ... | | Authors: | Uenaka, M, Ohnishi, Y, Tanaka, H, Kurisu, G. | | Deposit date: | 2024-11-13 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-12 | | Method: | NEUTRON DIFFRACTION (1.8 Å) | | Cite: | Redox-dependent hydrogen-bond network rearrangement of ferredoxin-NADP + reductase revealed by high-resolution X-ray and neutron crystallography.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
9KKG
 
 | | High resolution structure of Ferredoxin-NADP+ reductase from maize root - Oxidized form, low X-ray dose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, ... | | Authors: | Uenaka, M, Ohnishi, Y, Tanaka, H, Kurisu, G. | | Deposit date: | 2024-11-13 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Redox-dependent hydrogen-bond network rearrangement of ferredoxin-NADP + reductase revealed by high-resolution X-ray and neutron crystallography.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
7UBU
 
 | |
5UG7
 
 | | Calcium bound Perforin C2 Domain - T431D | | Descriptor: | CALCIUM ION, Perforin-1 | | Authors: | Law, R.H.P, Conroy, P.J, Voskoboinik, I, Whisstock, J.C. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Perforin proteostasis is regulated through its C2 domain: supra-physiological cell death mediated by T431D-perforin.
Cell Death Differ., 25, 2018
|
|
1X7I
 
 | | Crystal structure of the native copper homeostasis protein (cutCm) with calcium binding from Shigella flexneri 2a str. 301 | | Descriptor: | CALCIUM ION, Copper homeostasis protein cutC | | Authors: | Zhu, D.Y, Zhu, Y.Q, Huang, R.H, Xiang, Y, Wang, D.C. | | Deposit date: | 2004-08-14 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the copper homeostasis protein (CutCm) from Shigella flexneri at 1.7 A resolution: The first structure of a new sequence family of TIM barrels
Proteins, 58, 2004
|
|
7DWM
 
 | | Crystal structure of the phage VqmA-DPO complex | | Descriptor: | 3,5-dimethylpyrazin-2-ol, Transcriptional regulator | | Authors: | Gu, Y, Yang, W.S. | | Deposit date: | 2021-01-17 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Understanding the mechanism of asymmetric gene regulation determined by the VqmA of vibriophage.
Biochem.Biophys.Res.Commun., 558, 2021
|
|
5EPQ
 
 | | Structure at 1.75 A resolution of a glycosylated, lipid-binding, lipocalin-like protein | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Banerjee, S, Chavas, L.M.G, Ramaswamy, S. | | Deposit date: | 2015-11-12 | | Release date: | 2015-12-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Structure of a heterogeneous, glycosylated, lipid-bound, in vivo-grown protein crystal at atomic resolution from the viviparous cockroach Diploptera punctata.
Iucrj, 3, 2016
|
|
5IST
 
 | | Staphylococcus aureus Dihydrofolate Reductase complexed with beta-NADPH, cyclic alpha-NADPH anomer and 3'-(3-(2,4-diamino-6-ethylpyrimidin-5-yl)prop-2-yn-1-yl)-4'-methoxy-[1,1'-biphenyl]-4-carboxylic acid (UCP1106) | | Descriptor: | 4-[3-[3-[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]prop-2-ynyl]-4-methoxy-phenyl]benzoic acid, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Anderson, A.C, Reeve, S.M. | | Deposit date: | 2016-03-15 | | Release date: | 2017-06-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.723 Å) | | Cite: | Charged Propargyl-Linked Antifolates Reveal Mechanisms of Antifolate Resistance and Inhibit Trimethoprim-Resistant MRSA Strains Possessing Clinically Relevant Mutations.
J. Med. Chem., 59, 2016
|
|
5ISP
 
 | | Staphylococcus aureus F98Y Dihydrofolate Reductase mutant complexed with beta-NADPH and 3'-(3-(2,4-diamino-6-ethylpyrimidin-5-yl)prop-2-yn-1-yl)-4'-methoxy-[1,1'-biphenyl]-4-carboxylic acid (UCP1106) | | Descriptor: | 4-[3-[3-[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]prop-2-ynyl]-4-methoxy-phenyl]benzoic acid, Dihydrofolate reductase, GLYCEROL, ... | | Authors: | Anderson, A.C, Reeve, S.M. | | Deposit date: | 2016-03-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Charged Propargyl-Linked Antifolates Reveal Mechanisms of Antifolate Resistance and Inhibit Trimethoprim-Resistant MRSA Strains Possessing Clinically Relevant Mutations.
J. Med. Chem., 59, 2016
|
|
