4FJ4
 
 | | Crystal structure of the protein Q9HRE7 complexed with mercury from Halobacterium salinarium at the resolution 2.1A, Northeast Structural Genomics Consortium target HsR50 | | Descriptor: | ETHYL MERCURY ION, SODIUM ION, Uncharacterized protein | | Authors: | Kuzin, A, Chen, Y, Vorobiev, S.M, Seetharaman, J, Janjua, J, Xiao, R, Cunningham, K, Maglaqui, M, Owens, L.A, Wang, H, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-11 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Northeast Structural Genomics Consortium Target HsR50
To be Published
|
|
4F9K
 
 | | Crystal Structure of human cAMP-dependent protein kinase type I-beta regulatory subunit (fragment 11-73), Northeast Structural Genomics Consortium (NESG) Target HR8613A | | Descriptor: | cAMP-dependent protein kinase type I-beta regulatory subunit | | Authors: | Kuzin, A, Lew, S, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Shastry, R, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-05-18 | | Release date: | 2012-06-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of human cAMP-dependent protein kinase type I-beta regulatory subunit (fragment 11-73), Northeast Structural Genomics Consortium (NESG) Target HR8613A
To be Published
|
|
4FCZ
 
 | | Crystal Structure of Toluene-tolerance protein from Pseudomonas putida (strain KT2440), Northeast Structural Genomics Consortium (NESG) Target PpR99 | | Descriptor: | Toluene-tolerance protein | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-05-25 | | Release date: | 2013-02-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Northeast Structural Genomics Consortium Target PpR99
To be Published
|
|
4G06
 
 | | Crystal structure of protein SP_0782 (7-79) from Streptococcus pneumoniae complexed with ssDNA. Northeast Structural Genomics Consortium (NESG) target SPR104 | | Descriptor: | DI(HYDROXYETHYL)ETHER, THYMIDINE-5'-PHOSPHATE, Uncharacterized protein | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-07-09 | | Release date: | 2012-07-25 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.905 Å) | | Cite: | Northeast Structural Genomics Consortium Target SpR104
To be Published
|
|
4DIU
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR94 | | Descriptor: | Engineered Protein PF00326 | | Authors: | Seetharaman, J, Lew, S, Wang, D, Kohan, E, Patel, D, Whitehead, T, Fleishman, S, Ciccosanti, C, Xiao, R, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-01-31 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR94
To be Published
|
|
4DMB
 
 | | X-ray structure of human hepatitus C virus NS5A-transactivated protein 2 at the resolution 1.9A, Northeast Structural Genomics Consortium (NESG) Target HR6723 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, HD domain-containing protein 2, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2012-02-07 | | Release date: | 2012-04-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Northeast Structural Genomics Consortium Target HR6723
To be Published
|
|
4EVW
 
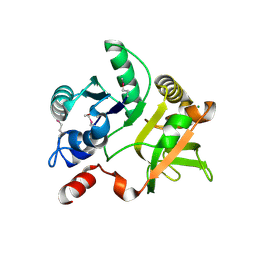 | | Crystal Structure of the nucleoside-diphosphate-sugar pyrophosphorylase from Vibrio cholerae RC9. Northeast Structural Genomics Consortium (NESG) Target VcR193. | | Descriptor: | MAGNESIUM ION, Nucleoside-diphosphate-sugar pyrophosphorylase | | Authors: | Vorobiev, S, Neely, H, Su, M, Seetharaman, J, Mao, M, Xiao, R, Kohan, E, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-26 | | Release date: | 2012-05-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the nucleoside-diphosphate-sugar pyrophosphorylase from Vibrio cholerae RC9.
To be Published
|
|
4GIF
 
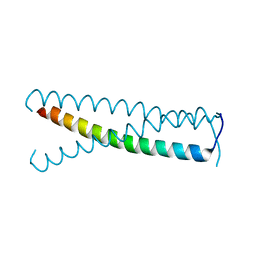 | | C-terminal coiled-coil domain of transient receptor potential channel TRPP3 (PKD2L1, Polycystin-L) | | Descriptor: | Polycystic kidney disease 2-like 1 protein | | Authors: | Yu, Y, Ulbrich, M.H, Li, M.-H, Dobbins, S, Zhang, W.K, Tong, L, Isacoff, E.Y, Yang, J. | | Deposit date: | 2012-08-08 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular mechanism of the assembly of an acid-sensing receptor ion channel complex.
Nat Commun, 3, 2012
|
|
4GPS
 
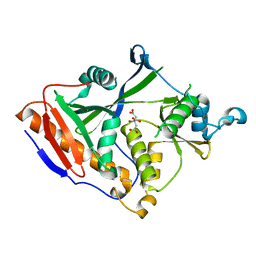 | |
4GPU
 
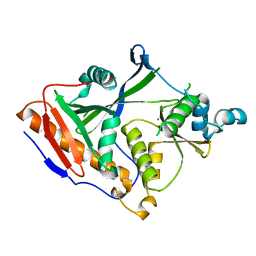 | |
4GVV
 
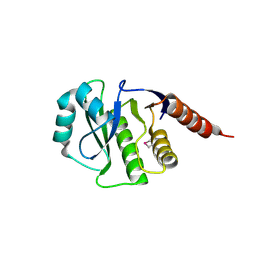 | | Crystal Structure of de novo design serine hydrolase OSH55.27, Northeast Structural Genomics Consortium (NESG) Target OR246 | | Descriptor: | De novo design serine hydrolase | | Authors: | Kuzin, A, Lew, S, Seetharaman, J, Mao, M, Xiao, R, Kohan, E, Rajagopalan, S, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-08-31 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.895 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR246
To be Published
|
|
4HFX
 
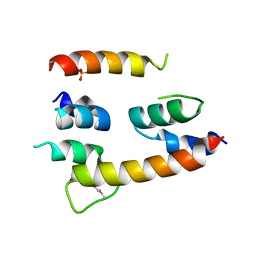 | | Crystal structure of a transcription elongation factor B polypeptide 3 from Homo sapiens, Northeast Structural Genomics consortium target id HR4748B. | | Descriptor: | SULFATE ION, Transcription elongation factor B polypeptide 3 | | Authors: | Seetharaman, J, Su, M, Ciccosanti, C, Sahdev, S, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-10-05 | | Release date: | 2012-12-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structure of a transcription elongation factor B polypeptide 3 from Homo sapiens, Northeast Structural Genomics consortium target id HR4748B. (CASP Target)
TO BE PUBLISHED
|
|
4H3H
 
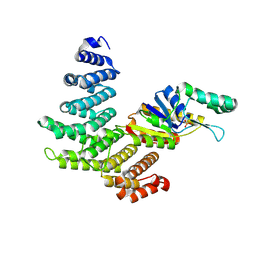 | |
4E6H
 
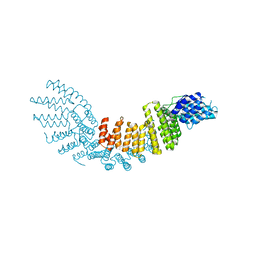 | |
4GVW
 
 | | Three-dimensional structure of the de novo designed serine hydrolase 2bfq_3, Northeast Structural Genomics Consortium (NESG) Target OR248 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETIC ACID, De novo designed serine hydrolase, ... | | Authors: | Kuzin, A, Lew, S, Seetharaman, J, Rajagopalan, S, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-08-31 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR248
To be Published
|
|
4HG0
 
 | | Crystal Structure of magnesium and cobalt efflux protein CorC, Northeast Structural Genomics Consortium (NESG) Target ER40 | | Descriptor: | ADENOSINE MONOPHOSPHATE, Magnesium and cobalt efflux protein CorC | | Authors: | Kuzin, A, Neely, H, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Cooper, B, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-10-05 | | Release date: | 2013-02-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Northeast Structural Genomics Consortium Target ER40
To be Published
|
|
2DLB
 
 | | X-ray Crystal Structure of Protein yopT from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR412 | | Descriptor: | yopT | | Authors: | Kuzin, A.P, Chen, Y, Seetharaman, J, Ho, C.-K, Cunningham, K, Janjua, H, Conover, K, Ma, L.-C, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-04-18 | | Release date: | 2006-04-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | X-ray structure of hypothetical protein from Bacillus subtilis O34498 at the resolution of 1.2A. NESG target SR412
To be published
|
|
2ELA
 
 | | Crystal Structure of the PTB domain of human APPL1 | | Descriptor: | Adapter protein containing PH domain, PTB domain and leucine zipper motif 1 | | Authors: | Li, J, Mao, X, Dong, L.Q, Liu, F, Tong, L. | | Deposit date: | 2007-03-27 | | Release date: | 2007-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the BAR-PH and PTB Domains of Human APPL1
Structure, 15, 2007
|
|
2ELB
 
 | | Crystal Structure of the BAR-PH domain of human APPL1 | | Descriptor: | Adapter protein containing PH domain, PTB domain and leucine zipper motif 1 | | Authors: | Li, J, Mao, X, Dong, L.Q, Liu, F, Tong, L. | | Deposit date: | 2007-03-27 | | Release date: | 2007-05-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the BAR-PH and PTB Domains of Human APPL1
Structure, 15, 2007
|
|
2ES9
 
 | | Crystal structure of Q8ZRJ2 from salmonella typhimurium. NESG TARGET STR65 | | Descriptor: | putative cytoplasmic protein | | Authors: | Benach, J, Abashidze, M, Jayaraman, S, Janjua, H, Cooper, B, Rong, X, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-10-25 | | Release date: | 2005-11-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Q8ZRJ2 from Salmonella typhimurium NESG TARGET STR65.
To be Published
|
|
2FFI
 
 | | Crystal Structure of Putative 2-Pyrone-4,6-Dicarboxylic Acid Hydrolase from Pseudomonas putida, Northeast Structural Genomics Target PpR23. | | Descriptor: | 2-pyrone-4,6-dicarboxylic acid hydrolase, putative, PHOSPHATE ION | | Authors: | Forouhar, F, Su, M, Jayaraman, S, Conover, K, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-19 | | Release date: | 2005-12-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structure of Putative 2-Pyrone-4,6-Dicarboxylic Acid Hydrolase from Pseudomonas putida, Northeast Structural Genomics Target PpR23.
To be Published
|
|
2GH1
 
 | | Crystal Structure of the putative SAM-dependent methyltransferase BC2162 from Bacillus cereus, Northeast Structural Genomics Target BcR20. | | Descriptor: | ACETATE ION, GLYCEROL, Methyltransferase | | Authors: | Forouhar, F, Neely, H, Jayaraman, S, Ciao, M, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-03-24 | | Release date: | 2006-04-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the putative SAM-dependent methyltransferase BC2162 from Bacillus cereus, Northeast Structural Genomics Target BcR20.
To be Published
|
|
2F9F
 
 | | Crystal Structure of the Putative Mannosyl Transferase (wbaZ-1)from Archaeoglobus fulgidus, Northeast Structural Genomics Target GR29A. | | Descriptor: | first mannosyl transferase (wbaZ-1) | | Authors: | Zhou, W, Forouhar, F, Conover, K, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-05 | | Release date: | 2006-06-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Putative Mannosyl Transferase
(wbaZ-1)from Archaeoglobus fulgidus
To be Published
|
|
2EWO
 
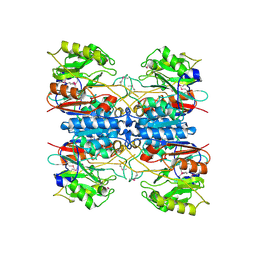 | |
2FFM
 
 | | X-Ray Crystal Structure of Protein SAV1430 from Staphylococcus aureus. Northeast Structural Genomics Consortium Target ZR18. | | Descriptor: | SAV1430 | | Authors: | Forouhar, F, Chen, Y, Jayaraman, S, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-19 | | Release date: | 2005-12-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal Structure of the Hypothetical Protein SAV1430 from Staphylococcus aureus, Northeast Structural Genomics ZR18.
To be Published
|
|
