7TLD
 
 | |
7TLB
 
 | |
7TP7
 
 | |
7TLC
 
 | |
7THE
 
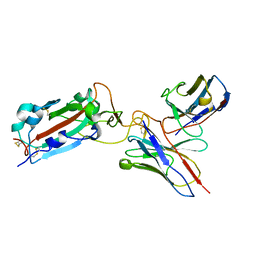 | | Structure of RBD directed antibody DH1042 in complex with SARS-CoV-2 spike: Local refinement of RBD-Fab interface | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH1042 Fab Heavy Chain, DH1042 Fab Light Chain, ... | | Authors: | May, A.J, Manne, K, Acharya, P. | | Deposit date: | 2022-01-10 | | Release date: | 2022-02-16 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structural diversity of the SARS-CoV-2 Omicron spike.
Mol.Cell, 82, 2022
|
|
7TOW
 
 | |
4WNX
 
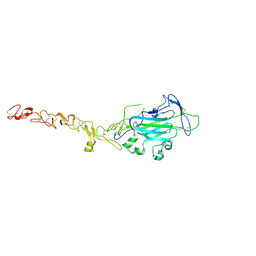 | | Netrin 4 lacking the C-terminal Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | McDougall, M, Patel, T, Reuten, R, Meier, M, Koch, M, Stetefeld, J. | | Deposit date: | 2014-10-14 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.723 Å) | | Cite: | Structural decoding of netrin-4 reveals a regulatory function towards mature basement membranes.
Nat Commun, 7, 2016
|
|
8K5W
 
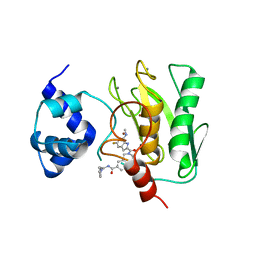 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | Descriptor: | 2-[[5-fluoranyl-7-(methylamino)-1H-indol-2-yl]carbonyl]-N-(2-pyrrol-1-ylethyl)-3,4-dihydro-1H-isoquinoline-7-carboxamide, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
8K5V
 
 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | Descriptor: | 6,7-dihydro-4H-[1,3]oxazolo[4,5-c]pyridin-5-yl-(7-ethyl-2H-indazol-3-yl)methanone, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
8K5Y
 
 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | Descriptor: | (3-azanyl-4-fluoranyl-5,7,8,9-tetrahydropyrido[4,3-c]azepin-6-yl)-[6-(2-oxidanylpropan-2-yl)-1H-indol-2-yl]methanone, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
8K5X
 
 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | Descriptor: | (6-cyclopropyl-1~{H}-indol-2-yl)-(5,7,8,9-tetrahydropyrido[4,3-c]azepin-6-yl)methanone, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
7PDJ
 
 | | R12E vFLIP mutant | | Descriptor: | FLICE inhibitory protein | | Authors: | Barrett, T.E. | | Deposit date: | 2021-08-05 | | Release date: | 2022-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Mechanistic insights into the activation of the IKK kinase complex by the Kaposi's sarcoma herpes virus oncoprotein vFLIP.
J.Biol.Chem., 298, 2022
|
|
7B83
 
 | | Structure of SARS-CoV-2 Main Protease bound to pyrithione zinc | | Descriptor: | 3C-like proteinase, 9-oxa-7-thia-1-azonia-8$l^{2}-zincabicyclo[4.3.0]nona-1,3,5-triene, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7UB0
 
 | |
7UB5
 
 | |
7UB6
 
 | |
5NUU
 
 | | Torpedo californica acetylcholinesterase in complex with a chlorotacrine-tryptophan hybrid inhibitor | | Descriptor: | (2~{S})-2-azanyl-~{N}-[6-[(6-chloranyl-1,2,3,4-tetrahydroacridin-9-yl)amino]hexyl]-3-(1~{H}-indol-3-yl)propanamide, Acetylcholinesterase, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Caliandro, R, Pesaresi, A, Lamba, D. | | Deposit date: | 2017-05-02 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel tacrine-tryptophan hybrids: Multi-target directed ligands as potential treatment for Alzheimer's disease.
Eur.J.Med.Chem., 168, 2019
|
|
5LDE
 
 | | Crystal structure of a vFLIP-IKKgamma stapled peptide dimer | | Descriptor: | Immunoglobulin G-binding protein G,Viral FLICE protein, Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma, ... | | Authors: | Barrett, T. | | Deposit date: | 2016-06-24 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | IKK gamma-Mimetic Peptides Block the Resistance to Apoptosis Associated with Kaposi's Sarcoma-Associated Herpesvirus Infection.
J. Virol., 91, 2017
|
|
7AQE
 
 | | Structure of SARS-CoV-2 Main Protease bound to UNC-2327 | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Meents, A. | | Deposit date: | 2020-10-21 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6WKU
 
 | | Twelve Chloride Ions Drive Assembly of Human alpha345 Collagen IV NC1 domain | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CHLORIDE ION, ... | | Authors: | Boudko, S.P, Hudson, B.G. | | Deposit date: | 2020-04-17 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Collagen IV alpha 345 dysfunction in glomerular basement membrane diseases. II. Crystal structure of the alpha 345 hexamer.
J.Biol.Chem., 296, 2021
|
|
7QT2
 
 | | Antibody FenAb208 - fentanyl complex | | Descriptor: | Antibody heavy chain, Antibody light chain, N-phenyl-N-[1-(2-phenylethyl)piperidin-4-yl]propanamide | | Authors: | Zeelen, J.P, Straaten van, M, Stebbins, C.E. | | Deposit date: | 2022-01-14 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A trypanosome-derived immunotherapeutics platform elicits potent high-affinity antibodies, negating the effects of the synthetic opioid fentanyl.
Cell Rep, 42, 2023
|
|
7QT0
 
 | | Antibody FenAb136 - fentanyl complex | | Descriptor: | 2-[2-[2-[2-[[5-oxidanylidene-5-[2-[4-[phenyl(propanoyl)amino]piperidin-1-yl]ethylamino]pentanoyl]amino]ethanoylamino]ethanoylamino]ethanoylamino]ethanoic acid, Antibody heavy chain, Antibody light chain | | Authors: | Zeelen, J.P, Straaten van, M, Stebbins, C.E. | | Deposit date: | 2022-01-14 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | A trypanosome-derived immunotherapeutics platform elicits potent high-affinity antibodies, negating the effects of the synthetic opioid fentanyl.
Cell Rep, 42, 2023
|
|
7QT4
 
 | | Antibody FenAb709 - fentanyl complex | | Descriptor: | 2-[2-[2-[2-[[5-oxidanylidene-5-[2-[4-[phenyl(propanoyl)amino]piperidin-1-yl]ethylamino]pentanoyl]amino]ethanoylamino]ethanoylamino]ethanoylamino]ethanoic acid, Antibody heavy chain, Antibody light chain | | Authors: | Zeelen, J.P, Straaten van, M, Stebbins, C.E. | | Deposit date: | 2022-01-14 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A trypanosome-derived immunotherapeutics platform elicits potent high-affinity antibodies, negating the effects of the synthetic opioid fentanyl.
Cell Rep, 42, 2023
|
|
7QT3
 
 | | Antibody FenAb609 - fentanyl complex | | Descriptor: | Antibody heavy chain, Antibody light chain, N-phenyl-N-[1-(2-phenylethyl)piperidin-4-yl]propanamide | | Authors: | Zeelen, J.P, Straaten van, M, Stebbins, C.E. | | Deposit date: | 2022-01-14 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A trypanosome-derived immunotherapeutics platform elicits potent high-affinity antibodies, negating the effects of the synthetic opioid fentanyl.
Cell Rep, 42, 2023
|
|
6V9C
 
 | | Crystal structure of FGFR4 kinase domain in complex with covalent inhibitor | | Descriptor: | Fibroblast growth factor receptor 4, N-[(3R,4S)-4-{[6-(2,6-dichloro-3,5-dimethoxyphenyl)-8-methyl-7-oxo-7,8-dihydropyrido[2,3-d]pyrimidin-2-yl]amino}oxolan-3-yl]prop-2-enamide, SULFATE ION | | Authors: | Liu, J, Liu, H. | | Deposit date: | 2019-12-13 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Selective, Covalent FGFR4 Inhibitors with Antitumor Activity in Models of Hepatocellular Carcinoma.
Acs Med.Chem.Lett., 11, 2020
|
|
