7MSM
 
 | | Mtb 70SIC in complex with MtbEttA at Trans_R0 state | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Cui, Z, Zhang, J. | | Deposit date: | 2021-05-11 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Interplay between an ATP-binding cassette F protein and the ribosome from Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
7MT2
 
 | | Mtb 70S initiation complex | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Cui, Z, Zhang, J. | | Deposit date: | 2021-05-12 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Interplay between an ATP-binding cassette F protein and the ribosome from Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
2OIG
 
 | | Crystal structure of RS21-C6 core segment and dm5CTP complex | | Descriptor: | 2'-DEOXY-5-METHYLCYTIDINE 5'-(TETRAHYDROGEN TRIPHOSPHATE), RS21-C6 | | Authors: | Wu, B, Liu, Y, Zhao, Q, Liao, S, Zhang, J, Bartlam, M, Chen, W, Rao, Z. | | Deposit date: | 2007-01-11 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of RS21-C6, Involved in Nucleoside Triphosphate Pyrophosphohydrolysis
J.Mol.Biol., 367, 2007
|
|
2OIE
 
 | | Crystal structure of RS21-C6 core segment RSCUT | | Descriptor: | RS21-C6, SULFATE ION | | Authors: | Wu, B, Liu, Y, Zhao, Q, Liao, S, Zhang, J, Bartlam, M, Chen, W, Rao, Z. | | Deposit date: | 2007-01-10 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of RS21-C6, Involved in Nucleoside Triphosphate Pyrophosphohydrolysis
J.Mol.Biol., 367, 2007
|
|
5GO0
 
 | |
5HNK
 
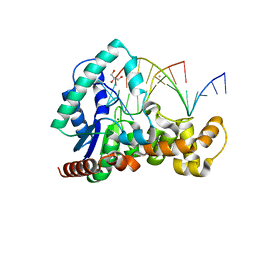 | | Crystal structure of T5Fen in complex intact substrate and metal ions. | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*GP*CP*GP*TP*AP*CP*GP*C)-3'), Exodeoxyribonuclease, GLYCEROL, ... | | Authors: | Almalki, F.A, Feng, M, Zhang, J, Sedelnikova, S.E, Rafferty, J.B, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2016-01-18 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Direct observation of DNA threading in flap endonuclease complexes.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5HRB
 
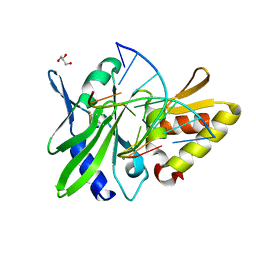 | | The crystal structure of AsfvPolX:DNA1 binary complex | | Descriptor: | BETA-MERCAPTOETHANOL, DNA (5'-D(*CP*GP*GP*AP*TP*AP*TP*CP*C)-3'), DNA polymerase beta-like protein, ... | | Authors: | Chen, Y.Q, Zhang, J, Gan, J.H. | | Deposit date: | 2016-01-23 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of Se-AsfvPolX(L52/163M mutant) in complex with 1nt-gap DNA1
To Be Published
|
|
5HRH
 
 | |
5HRD
 
 | |
3HAG
 
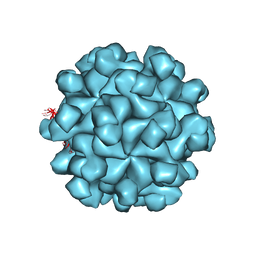 | | Crystal structure of the Hepatitis E Virus-like Particle | | Descriptor: | Capsid protein | | Authors: | Guu, T.S.Y, Liu, Z, Ye, Q, Mata, D.A, Li, K, Yin, C, Zhang, J, Tao, Y.J. | | Deposit date: | 2009-05-01 | | Release date: | 2009-09-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the hepatitis E virus-like particle suggests mechanisms for virus assembly and receptor binding.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7C2Q
 
 | | The crystal structure of COVID-19 main protease in the apo state | | Descriptor: | 3C-like proteinase | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Hu, X.H, Zhou, H, Wang, Q.S, Li, j, Zhang, J. | | Deposit date: | 2020-05-08 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of SARS-CoV-2 main protease in the apo state.
Sci China Life Sci, 64, 2021
|
|
2KLI
 
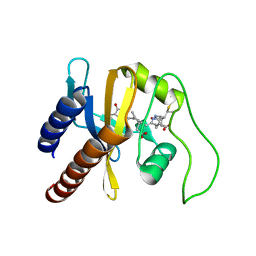 | | Structural Basis for the Photoconversion of A Phytochrome to the Activated FAR-RED LIGHT-ABSORBING Form | | Descriptor: | PHYCOCYANOBILIN, Sensor protein | | Authors: | Cornilescu, C.C, Cornilescu, G, Ulijasz, A.T, Zhang, J, Rivera, M, Vierstra, R.D, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-07-03 | | Release date: | 2009-11-03 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the photoconversion of a phytochrome to the activated Pfr form
Nature, 463, 2010
|
|
2ID0
 
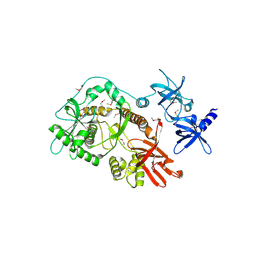 | | Escherichia coli RNase II | | Descriptor: | Exoribonuclease 2, MANGANESE (II) ION | | Authors: | Zuo, Y, Zhang, J, Wang, Y, Malhotra, A. | | Deposit date: | 2006-09-13 | | Release date: | 2006-10-03 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for Processivity and Single-Strand Specificity of RNase II.
Mol.Cell, 24, 2006
|
|
7CA8
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Li, J, Zhang, J. | | Deposit date: | 2020-06-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease in complex with the natural product inhibitor shikonin illuminates a unique binding mode.
Sci Bull (Beijing), 66, 2021
|
|
7XJF
 
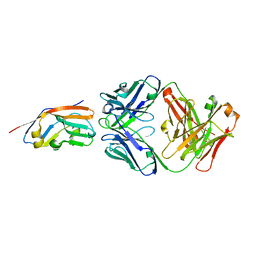 | | Crystal structure of 6MW3211 Fab in complex with CD47 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Wang, J, Wang, R, Jiao, S, Wang, S, Zhang, J, Zhang, M, Wang, M. | | Deposit date: | 2022-04-16 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Blockade of dual immune checkpoint inhibitory signals with a CD47/PD-L1 bispecific antibody for cancer treatment.
Theranostics, 13, 2023
|
|
8TS2
 
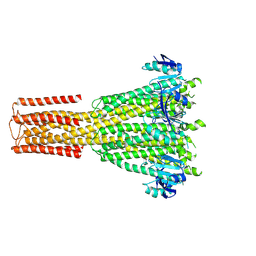 | | Cryo-EM structure of human MRS2 with EDTA | | Descriptor: | Magnesium transporter MRS2 homolog, mitochondrial, Soluble cytochrome b562 fusion protein | | Authors: | He, Z, Mount, J.W, Zhang, J, Yuan, P. | | Deposit date: | 2023-08-10 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Human mitochondrial MRS2 is a Ca2+-regulated nonselective cation channel
To Be Published
|
|
8TS3
 
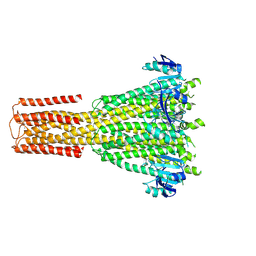 | | Cryo-EM structure of human MRS2 with Ca2+ | | Descriptor: | CALCIUM ION, Magnesium transporter MRS2 homolog, mitochondrial, ... | | Authors: | He, Z, Mount, J.W, Zhang, J, Yuan, P. | | Deposit date: | 2023-08-10 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Human mitochondrial MRS2 is a Ca2+-regulated nonselective cation channel
To Be Published
|
|
8TS1
 
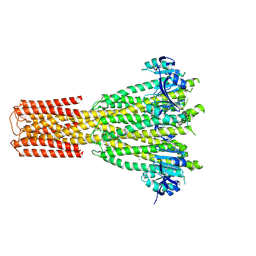 | | Cryo-EM structure of human MRS2 with Mg2+ | | Descriptor: | MAGNESIUM ION, Magnesium transporter MRS2 homolog, mitochondrial, ... | | Authors: | He, Z, Mount, J.W, Zhang, J, Yuan, P. | | Deposit date: | 2023-08-10 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Human mitochondrial MRS2 is a Ca2+-regulated nonselective cation channel
To Be Published
|
|
7DAC
 
 | | Human RIPK3 amyloid fibril revealed by solid-state NMR | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Wu, X.L, Zhang, J, Dong, X.Q, Liu, J, Li, B, Hu, H, Wang, J, Wang, H.Y, Lu, J.X. | | Deposit date: | 2020-10-16 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | The structure of a minimum amyloid fibril core formed by necroptosis-mediating RHIM of human RIPK3.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DK0
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with MW05 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MW05 heavy chain, MW05 light chain, ... | | Authors: | Wang, J, Jiao, S, Wang, R, Zhang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-11-22 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.199 Å) | | Cite: | Antibody-dependent enhancement (ADE) of SARS-CoV-2 pseudoviral infection requires Fc gamma RIIB and virus-antibody complex with bivalent interaction.
Commun Biol, 5, 2022
|
|
2H7B
 
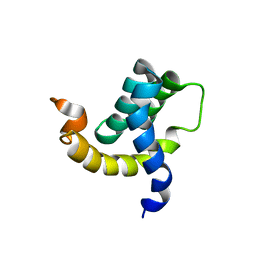 | | Solution structure of the eTAFH domain from the human leukemia-associated fusion protein AML1-ETO | | Descriptor: | Core-binding factor, ML1-ETO | | Authors: | Plevin, M.J, Zhang, J, Guo, C, Roeder, R.G, Ikura, M. | | Deposit date: | 2006-06-01 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The acute myeloid leukemia fusion protein AML1-ETO targets E proteins via a paired amphipathic helix-like TBP-associated factor homology domain
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2L7Y
 
 | |
2LJ8
 
 | |
7DYS
 
 | |
2LRW
 
 | | Solution structure of a ubiquitin-like protein from Trypanosoma brucei | | Descriptor: | Ubiquitin, putative | | Authors: | Wang, R, Wang, T, Liao, S, Zhang, J, Tu, X. | | Deposit date: | 2012-04-13 | | Release date: | 2013-04-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a ubiqutin-like protein from Trypanosoma bucei
To be Published
|
|
