2QKC
 
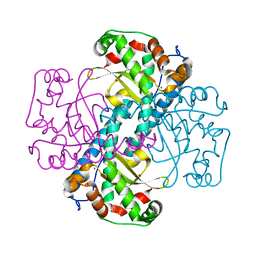 | | Structural and Kinetic Study of the Differences between Human and E.coli Manganese Superoxide Dismutases | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn] | | Authors: | Zheng, J, Domsic, J.F, Cabelli, D, McKenna, R, Silverman, D.N. | | Deposit date: | 2007-07-10 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic study of differences between human and Escherichia coli manganese superoxide dismutases.
Biochemistry, 46, 2007
|
|
7VWA
 
 | |
7VW8
 
 | |
7VW9
 
 | |
7X2P
 
 | |
3GBI
 
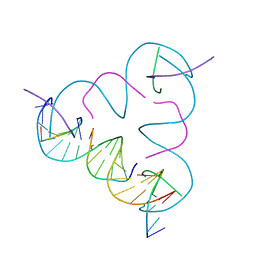 | | The Rational Design and Structural Analysis of a Self-Assembled Three-Dimensional DNA Crystal | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), ... | | Authors: | Birktoft, J.J, Zheng, J, Seeman, N.C. | | Deposit date: | 2009-02-19 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.018 Å) | | Cite: | From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal.
Nature, 461, 2009
|
|
4J1S
 
 | |
4J1Q
 
 | |
5XDM
 
 | |
7X2R
 
 | |
7XFF
 
 | | Polysaccharide export protein Wza | | Descriptor: | Polysaccharide export protein | | Authors: | Zheng, J, Wang, B. | | Deposit date: | 2022-04-01 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Polysaccharide export protein Wza
To Be Published
|
|
7XFD
 
 | | Polysaccharide export protein Wza | | Descriptor: | Polysaccharide export protein | | Authors: | Zheng, J, Wang, B. | | Deposit date: | 2022-04-01 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | polysaccharide export protein Wza
To Be Published
|
|
3CIO
 
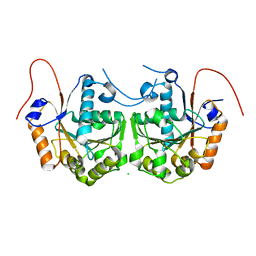 | |
6XMK
 
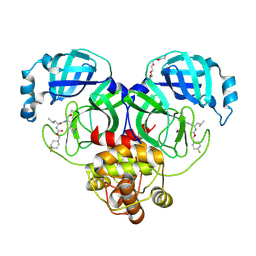 | | 1.70 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 7j | | Descriptor: | (1S,2S)-2-[(N-{[(4,4-difluorocyclohexyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
4V0R
 
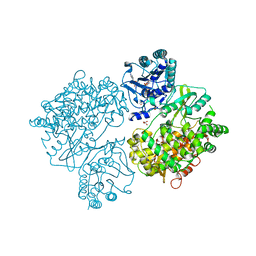 | | DENGUE VIRUS FULL LENGTH NS5 COMPLEXED WITH GTP AND SAH | | Descriptor: | FORMIC ACID, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhao, Y, Soh, S, Zheng, J, Phoo, W.W, Swaminathan, K, Cornvik, T.C, Lim, S.P, Shi, P.-Y, Lescar, J, Vasudevan, S.G, Luo, D. | | Deposit date: | 2014-09-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Crystal Structure of the Dengue Virus Ns5 Protein Reveals a Novel Inter-Domain Interface Essential for Protein Flexibility and Virus Replication.
Plos Pathog., 11, 2015
|
|
4V0Q
 
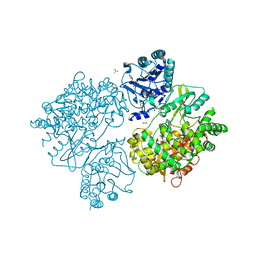 | | Dengue Virus Full Length NS5 Complexed with SAH | | Descriptor: | ACETATE ION, GLYCEROL, NS5 POLYMERASE, ... | | Authors: | Zhao, Y, Soh, S, Zheng, J, Phoo, W.W, Swaminathan, K, Cornvik, T.C, Lim, S.P, Shi, P.-Y, Lescar, J, Vasudevan, S.G, Luo, D. | | Deposit date: | 2014-09-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Crystal Structure of the Dengue Virus Ns5 Protein Reveals a Novel Inter-Domain Interface Essential for Protein Flexibility and Virus Replication.
Plos Pathog., 11, 2015
|
|
1CTP
 
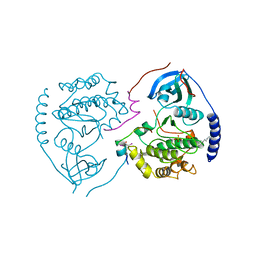 | | STRUCTURE OF THE MAMMALIAN CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE AND AN INHIBITOR PEPTIDE DISPLAYS AN OPEN CONFORMATION | | Descriptor: | MYRISTIC ACID, cAMP-DEPENDENT PROTEIN KINASE, cAMP-dependent protein kinase inhibitor, ... | | Authors: | Karlsson, R, Zheng, J, Xuong, N.H, Taylor, S.S, Sowadski, J.M. | | Deposit date: | 1993-04-08 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the mammalian catalytic subunit of cAMP-dependent protein kinase and an inhibitor peptide displays an open conformation.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
8IA9
 
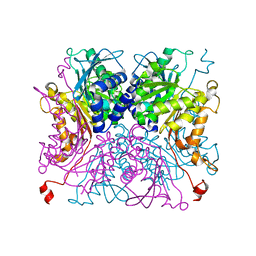 | |
8IAA
 
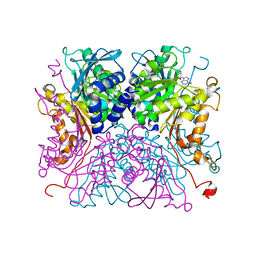 | |
1FSH
 
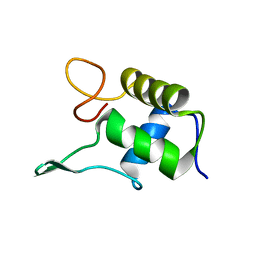 | |
1APM
 
 | | 2.0 ANGSTROM REFINED CRYSTAL STRUCTURE OF THE CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE COMPLEXED WITH A PEPTIDE INHIBITOR AND DETERGENT | | Descriptor: | N-OCTANE, PEPTIDE INHIBITOR PKI(5-24), cAMP-DEPENDENT PROTEIN KINASE | | Authors: | Knighton, D.R, Bell, S.M, Zheng, J, Teneyck, L.F, Xuong, N.-H, Taylor, S.S, Sowadski, J.M. | | Deposit date: | 1993-01-18 | | Release date: | 1993-04-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 A refined crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with a peptide inhibitor and detergent.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
6L3N
 
 | |
1NMI
 
 | | Solution structure of the imidazole complex of iso-1 cytochrome c | | Descriptor: | Cytochrome c, iso-1, HEME C, ... | | Authors: | Yao, Y, Tong, Y, Liu, G, Wang, J, Zheng, J, Tang, W. | | Deposit date: | 2003-01-10 | | Release date: | 2003-02-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the imidazole complex of iso-1 cytochrome c
To be Published
|
|
7DNJ
 
 | | K63-polyUb MDA5CARDs complex | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, Ubiquitin | | Authors: | Song, B, Chen, Y, Luo, D.H, Zheng, J. | | Deposit date: | 2020-12-09 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ordered assembly of the cytosolic RNA-sensing MDA5-MAVS signaling complex via binding to unanchored K63-linked poly-ubiquitin chains.
Immunity, 54, 2021
|
|
7DNI
 
 | | MDA5 CARDs-MAVS CARD polyUb complex | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, Mitochondrial antiviral-signaling protein, Ubiquitin | | Authors: | Song, B, Chen, Y, Luo, D.H, Zheng, J. | | Deposit date: | 2020-12-09 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Ordered assembly of the cytosolic RNA-sensing MDA5-MAVS signaling complex via binding to unanchored K63-linked poly-ubiquitin chains.
Immunity, 54, 2021
|
|
