8GNA
 
 | | Structure of the SbCas7-11-crRNA-NTR complex | | Descriptor: | RAMP superfamily protein, RNA (32-MER), RNA (5'-R(P*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*GP*GP*U)-3'), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-08-23 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Target RNA-guided protease activity in type III-E CRISPR-Cas system.
Nucleic Acids Res., 50, 2022
|
|
5WYZ
 
 | | Crystal structure of human TLR8 in complex with CU-CPT9b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(3-methyl-4-oxidanyl-phenyl)quinolin-7-ol, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2017-01-16 | | Release date: | 2017-12-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-molecule inhibition of TLR8 through stabilization of its resting state
Nat. Chem. Biol., 14, 2018
|
|
5WYX
 
 | | Crystal structure of human TLR8 in complex with CU-CPT8m | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-(3-methylphenyl)pyrazolo[1,5-a]pyrimidine-3-carboxamide, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2017-01-16 | | Release date: | 2017-12-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Small-molecule inhibition of TLR8 through stabilization of its resting state
Nat. Chem. Biol., 14, 2018
|
|
8Z9C
 
 | |
8Z4J
 
 | |
8Z4L
 
 | |
8Z9E
 
 | |
8Z99
 
 | |
8YHD
 
 | |
8YHE
 
 | |
7YUF
 
 | | apo human SPNS2 | | Descriptor: | NbFab H-chain, NbFab L-chain, Sphingosine-1-phosphate transporter SPNS2, ... | | Authors: | He, Y, Duan, Y. | | Deposit date: | 2022-08-17 | | Release date: | 2023-09-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural basis of Sphingosine-1-phosphate transport via human SPNS2.
Cell Res., 34, 2024
|
|
7YUD
 
 | | FTY720p-bound human SPNS2 | | Descriptor: | (2~{S})-2-azanyl-4-(4-octylphenyl)-2-[[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxymethyl]butan-1-ol, NbFab L-chain, NbFab-H-chain, ... | | Authors: | He, Y, Duan, Y. | | Deposit date: | 2022-08-17 | | Release date: | 2023-09-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of Sphingosine-1-phosphate transport via human SPNS2.
Cell Res., 34, 2024
|
|
4IST
 
 | |
4ISS
 
 | |
5TMA
 
 | | Zymomonas mobilis pyruvate decarboxylase mutant PDC-2.3 | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Pyruvate decarboxylase, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2016-10-12 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | An iterative computational design approach to increase the thermal endurance of a mesophilic enzyme.
Biotechnol Biofuels, 11, 2018
|
|
8KAE
 
 | | 16d-bound human SPNS2 | | Descriptor: | 3-[3-(4-decylphenyl)-1,2,4-oxadiazol-5-yl]propan-1-amine, NbFab chain L, NbFab-H chain, ... | | Authors: | He, Y, Duan, Y. | | Deposit date: | 2023-08-03 | | Release date: | 2024-01-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis of Sphingosine-1-phosphate transport via human SPNS2.
Cell Res., 34, 2024
|
|
5Z15
 
 | | Crystal structure of human TLR8 in complex with CU-CPT9c | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(7-chloranylquinolin-4-yl)-2-methyl-phenol, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2017-12-25 | | Release date: | 2018-07-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Inhibition of Toll-like Receptor 8 by Specifically Targeting a Unique Allosteric Site and Locking Its Resting State
To Be Published
|
|
5Z14
 
 | | Crystal structure of human TLR8 in complex with CU-CPT9a | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(7-methoxyquinolin-4-yl)-2-methyl-phenol, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2017-12-25 | | Release date: | 2018-07-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Inhibition of Toll-like Receptor 8 by Specifically Targeting a Unique Allosteric Site and Locking Its Resting State
To Be Published
|
|
5D06
 
 | |
5D0F
 
 | |
8WM9
 
 | | Fzd4/DEP complex | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Frizzled-4, Segment polarity protein dishevelled homolog DVL-2 | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2023-10-03 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural basis of Frizzled 4 in recognition of Dishevelled 2 unveils mechanism of WNT signaling activation.
Nat Commun, 15, 2024
|
|
8WMA
 
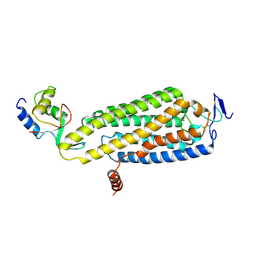 | | Fzd4/DEP complex (local refined) | | Descriptor: | Frizzled-4, Segment polarity protein dishevelled homolog DVL-2 | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2023-10-03 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural basis of Frizzled 4 in recognition of Dishevelled 2 unveils mechanism of WNT signaling activation.
Nat Commun, 15, 2024
|
|
5SUN
 
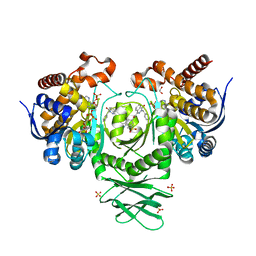 | | IDH1 R132H in complex with IDH146 | | Descriptor: | 1,2-ETHANEDIOL, 3-benzyl-N-[3-(dimethylsulfamoyl)phenyl]-4-oxo-3,4-dihydrophthalazine-1-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Xie, X, Kulathila, R. | | Deposit date: | 2016-08-03 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.477 Å) | | Cite: | Allosteric Mutant IDH1 Inhibitors Reveal Mechanisms for IDH1 Mutant and Isoform Selectivity.
Structure, 25, 2017
|
|
5SVF
 
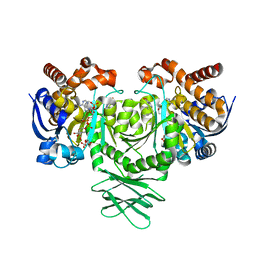 | | IDH1 R132H in complex with IDH125 | | Descriptor: | (4S)-3-(2-{[(1S)-1-phenylethyl]amino}pyrimidin-4-yl)-4-(propan-2-yl)-1,3-oxazolidin-2-one, CITRATE ANION, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Xie, X, Kulathila, R. | | Deposit date: | 2016-08-05 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Allosteric Mutant IDH1 Inhibitors Reveal Mechanisms for IDH1 Mutant and Isoform Selectivity.
Structure, 25, 2017
|
|
5SVO
 
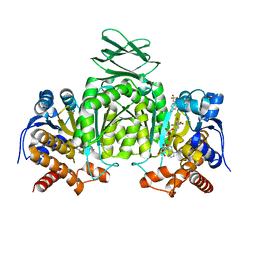 | | Structure of IDH2 mutant R140Q | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Isocitrate dehydrogenase [NADP], mitochondrial, ... | | Authors: | Xie, X, Kulathila, R. | | Deposit date: | 2016-08-06 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Allosteric Mutant IDH1 Inhibitors Reveal Mechanisms for IDH1 Mutant and Isoform Selectivity.
Structure, 25, 2017
|
|
