4GO1
 
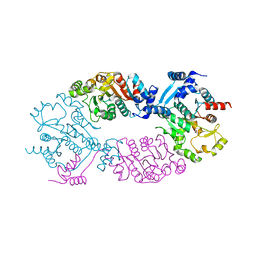 | | Crystal Structure of full length transcription repressor LsrR from E. coli. | | Descriptor: | GLYCEROL, Transcriptional regulator LsrR | | Authors: | Wu, M, Tao, Y, Liu, X, Zang, J. | | Deposit date: | 2012-08-17 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Phosphorylated Autoinducer-2 Modulation of the Oligomerization State of the Global Transcription Regulator LsrR from Escherichia coli
J.Biol.Chem., 288, 2013
|
|
1YHN
 
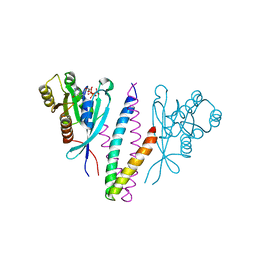 | | Structure basis of RILP recruitment by Rab7 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Rab interacting lysosomal protein, ... | | Authors: | Wu, M, Wang, T, Hong, W, Song, H. | | Deposit date: | 2005-01-10 | | Release date: | 2005-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for recruitment of RILP by small GTPase Rab7.
Embo J., 24, 2005
|
|
3RB7
 
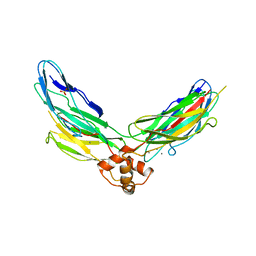 | | Crystal structure of CBD12 from CALX1.2 | | Descriptor: | CALCIUM ION, Na/Ca exchange protein, SULFATE ION | | Authors: | Wu, M, Zheng, L. | | Deposit date: | 2011-03-28 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis of the Ca(2+) Inhibitory Mechanism of Drosophila Na(+)/Ca(2+) Exchanger CALX and Its Modification by Alternative Splicing.
Structure, 19, 2011
|
|
3RB5
 
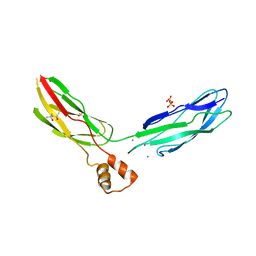 | | Crystal structure of calcium binding domain CBD12 of CALX1.1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Na/Ca exchange protein, ... | | Authors: | Wu, M, Zheng, L. | | Deposit date: | 2011-03-28 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of the Ca(2+) Inhibitory Mechanism of Drosophila Na(+)/Ca(2+) Exchanger CALX and Its Modification by Alternative Splicing.
Structure, 19, 2011
|
|
8ZH3
 
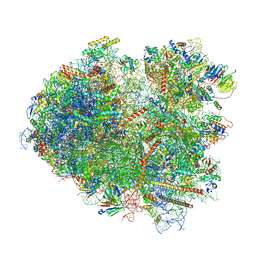 | | 80S ribosome with A/P-P/E tRNA and mRNA of WNV | | Descriptor: | 25S rRNA (3393-MER), 5.8S rRNA (158-MER), 5S rRNA (121-MER), ... | | Authors: | Wu, M, Yuan, S. | | Deposit date: | 2024-05-10 | | Release date: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Flexibilities and conservations in the structural basis of viral -1 programmed ribosomal frameshifting
To Be Published
|
|
6B72
 
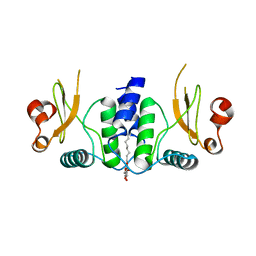 | | A novel HIV-1 Nef dimer interface induced by a single octyl-glucoside molecule | | Descriptor: | Protein Nef, octyl beta-D-glucopyranoside | | Authors: | Wu, M, Augelli-Szafran, C.E, Ptak, R.G, Smithgall, T.E. | | Deposit date: | 2017-10-03 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A single beta-octyl glucoside molecule induces HIV-1 Nef dimer formation in the absence of partner protein binding.
PLoS ONE, 13, 2018
|
|
7T3F
 
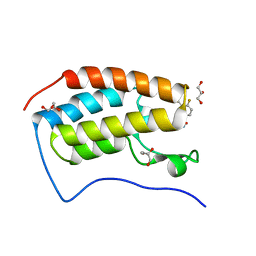 | | Development of BRD4 inhibitors as arsenicals antidotes | | Descriptor: | 4-fluoro-3-methyl-N-(3-methyl-2-oxo-1,2,3,4-tetrahydroquinazolin-6-yl)benzene-1-sulfonamide, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Wu, M, Yatchang, M, Mathew, B, Zhai, L, Ruiz, P, Bostwick, R, Augelli-Szafran, C.E, Suto, M.J. | | Deposit date: | 2021-12-07 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Development of BRD4 inhibitors as anti-inflammatory agents and antidotes for arsenicals.
Bioorg.Med.Chem.Lett., 64, 2022
|
|
5GUP
 
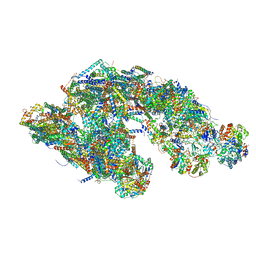 | | Cryo-EM structure of mammalian respiratory supercomplex I1III2IV1 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Acyl carrier protein, ... | | Authors: | Gu, J, Wu, M, Guo, R, Yang, M. | | Deposit date: | 2016-08-30 | | Release date: | 2017-03-29 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of Mammalian Respiratory Supercomplex I1III2IV1
Cell, 167, 2016
|
|
6VUQ
 
 | | Crystal structure of CHIKV nsP3 macrodomain soaked with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Nonstructural polyprotein | | Authors: | Wu, M. | | Deposit date: | 2020-02-16 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Pyrimidone inhibitors targeting Chikungunya Virus nsP3 macrodomain by fragment-based drug design.
Plos One, 16, 2021
|
|
6W0T
 
 | |
5WTA
 
 | | Crystal Structure of Staphylococcus aureus SdrE apo form | | Descriptor: | Serine-aspartate repeat-containing protein E | | Authors: | Wu, M, Zhang, Y, Hang, T, Wang, C, Yang, Y, Zang, J, Zhang, M, Zhang, X. | | Deposit date: | 2016-12-10 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Staphylococcus aureus SdrE captures complement factor H's C-terminus via a novel 'close, dock, lock and latch' mechanism for complement evasion
Biochem. J., 474, 2017
|
|
5WTB
 
 | | Complex Structure of Staphylococcus aureus SdrE with human complement factor H | | Descriptor: | Peptide from Complement factor H, Serine-aspartate repeat-containing protein E | | Authors: | Wu, M, Zhang, Y, Hang, T, Wang, C, Yang, Y, Zang, J, Zhang, M, Zhang, X. | | Deposit date: | 2016-12-10 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Staphylococcus aureus SdrE captures complement factor H's C-terminus via a novel 'close, dock, lock and latch' mechanism for complement evasion
Biochem. J., 474, 2017
|
|
3N2Y
 
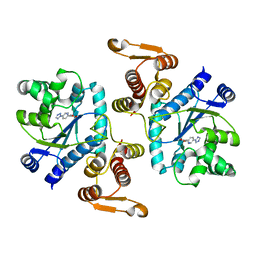 | |
1T91
 
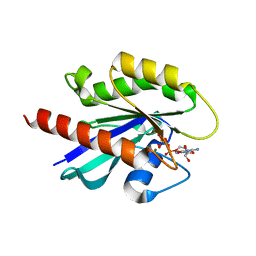 | | crystal structure of human small GTPase Rab7(GTP) | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-7 | | Authors: | Song, H, Hong, W, Wu, M, Wang, T. | | Deposit date: | 2004-05-14 | | Release date: | 2005-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for recruitment of RILP by small GTPase Rab7.
Embo J., 24, 2005
|
|
4E4H
 
 | | Crystal structure of Histone Demethylase NO66 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | Authors: | Wu, M, Tao, Y, Zang, J. | | Deposit date: | 2012-03-13 | | Release date: | 2013-01-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | tructural Basis for Tetramerization-dependent Gene Repression by Histone Demethylase NO66
To be Published
|
|
3P12
 
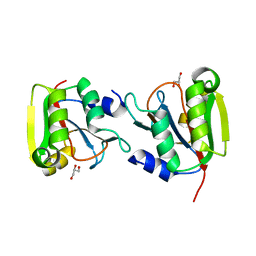 | | Crystal Structure of D-ribose Pyranase Sa240 | | Descriptor: | D-ribose pyranase, GLYCEROL | | Authors: | Wu, M, Wang, L, Zang, J. | | Deposit date: | 2010-09-29 | | Release date: | 2011-05-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of Sa240: A ribose pyranase homolog with partial active site from Staphylococcus aureus
J.Struct.Biol., 174, 2011
|
|
3P13
 
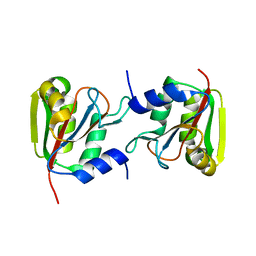 | |
4J0C
 
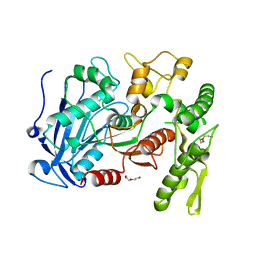 | | tannin acyl hydrolase from Lactobacillus plantarum (native structure) | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, Tannase | | Authors: | Wu, M, Wang, Q, Peng, X, Wen, H, Chen, Q, McKinstry, W.J. | | Deposit date: | 2013-01-30 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of tannase from Lactobacillus plantarum.
J.Mol.Biol., 425, 2013
|
|
5YCO
 
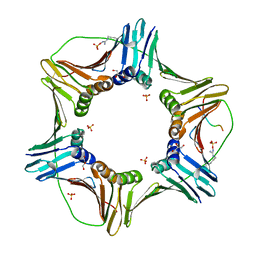 | | Complex structure of PCNA with UHRF2 | | Descriptor: | E3 ubiquitin-protein ligase UHRF2, GLYCEROL, Proliferating cell nuclear antigen, ... | | Authors: | Wu, M, Chen, W, Hang, T, Wang, C, Zhang, X, Zang, J. | | Deposit date: | 2017-09-07 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Structure insights into the molecular mechanism of the interaction between UHRF2 and PCNA.
Biochem. Biophys. Res. Commun., 494, 2017
|
|
6T8U
 
 | | Complement factor B in complex with 5-Bromo-3-chloro-N-(4,5-dihydro-1H-imidazol-2-yl)-7-methyl-1H-indol-4-amine | | Descriptor: | 5-bromanyl-3-chloranyl-~{N}-(1~{H}-imidazol-2-yl)-7-methyl-1~{H}-indol-4-amine, Complement factor B, SULFATE ION | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Mac Sweeney, A, Wiesmann, C, Adams, C, Liao, S.-M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Serrano-Wu, M, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, De Erkenez, A, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
6T8V
 
 | | Complement factor B in complex with (S)-5,7-Dimethyl-4-((2-phenylpiperidin-1-yl)methyl)-1H-indole | | Descriptor: | 4-[(2~{S})-1-[(5,7-dimethyl-1~{H}-indol-4-yl)methyl]piperidin-2-yl]benzoic acid, Complement factor B, SULFATE ION, ... | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Mac Sweeney, A, Wiesmann, C, Adams, C, Mainolfi, N, Liao, S.-M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Serrano-Wu, M, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, De Erkenez, A, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Wiesmann, C, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
8ZYB
 
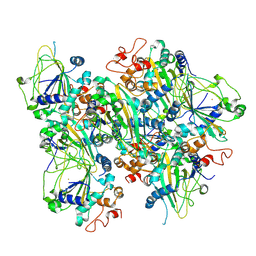 | |
2ISY
 
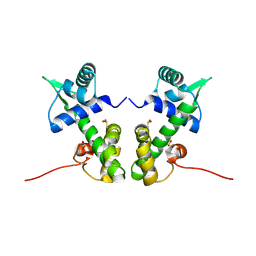 | | Crystal structure of the nickel-activated two-domain iron-dependent regulator (IdeR) | | Descriptor: | Iron-dependent repressor ideR, NICKEL (II) ION, PHOSPHATE ION | | Authors: | Wisedchaisri, G, Chou, C.J, Wu, M, Roach, C, Rice, A.E, Holmes, R.K, Beeson, C, Hol, W.G. | | Deposit date: | 2006-10-18 | | Release date: | 2007-02-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | Crystal structures, metal activation, and DNA-binding properties of two-domain IdeR from Mycobacterium tuberculosis
Biochemistry, 46, 2007
|
|
5XTI
 
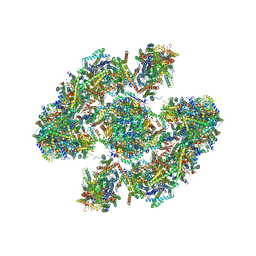 | | Cryo-EM architecture of human respiratory chain megacomplex-I2III2IV2 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Acyl carrier protein, ... | | Authors: | Gu, J, Wu, M, Yang, M. | | Deposit date: | 2017-06-19 | | Release date: | 2017-09-13 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (17.4 Å) | | Cite: | Architecture of Human Mitochondrial Respiratory Megacomplex I2III2IV2.
Cell, 170, 2017
|
|
5XTB
 
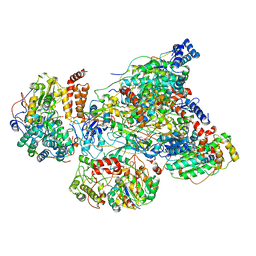 | | Cryo-EM structure of human respiratory complex I matrix arm | | Descriptor: | Acyl carrier protein, mitochondrial, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Gu, J, Wu, M, Yang, M. | | Deposit date: | 2017-06-18 | | Release date: | 2017-08-30 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Architecture of Human Mitochondrial Respiratory Megacomplex I2III2IV2.
Cell, 170, 2017
|
|
