4N1K
 
 | | Crystal structures of NLRP14 pyrin domain reveal a conformational switch mechanism, regulating its molecular interactions | | Descriptor: | NACHT, LRR and PYD domains-containing protein 14 | | Authors: | Eibl, C, Hessenberger, M, Wenger, J, Brandstetter, H. | | Deposit date: | 2013-10-04 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the NLRP14 pyrin domain reveal a conformational switch mechanism regulating its molecular interactions.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4N1J
 
 | | Crystal structures of NLRP14 pyrin domain reveal a conformational switch mechanism, regulating its molecular interactions | | Descriptor: | GLYCEROL, NACHT, LRR and PYD domains-containing protein 14 | | Authors: | Eibl, C, Hessenberger, M, Wenger, J, Brandstetter, H. | | Deposit date: | 2013-10-04 | | Release date: | 2014-07-16 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the NLRP14 pyrin domain reveal a conformational switch mechanism regulating its molecular interactions.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1OCI
 
 | | [3.2.0]bcANA:DNA | | Descriptor: | 5'-D(*CP*TP*GP*A TLBP*AP*TP*GP*CP)-3', 5'-D(*GP*CP*AP*TP*AP*TP*CP*AP*GP)-3' | | Authors: | Tommerholt, H.V, Christensen, N.K, Nielsen, P, Wengel, J, Stein, P.C, Jacobsen, J.P, Petersen, M. | | Deposit date: | 2003-02-07 | | Release date: | 2003-07-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of a DsDNA Containing a Bicyclic D-Arabino-Configured Nucleotide Fixed in an O4'-Endo Sugar Conformation
Org.Biomol.Chem., 1, 2003
|
|
1GV6
 
 | | Solution structure of alfa-L-LNA:DNA duplex | | Descriptor: | 5- D(*CP*(ATL)P*GP*CP*(ATL)P*(ATL)P*CP*(ATL)P* GP*C) -3, 5- D(*GP*CP*AP*GP*AP*AP*GP*CP*AP*G) -3 | | Authors: | Nielsen, K.M.E, Petersen, M, Haakansson, A.E, Wengel, J, Jacobsen, J.P. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Alfa-L-Lna (Alfa-L-Ribo Configured Locked Nucleic Acid) Recognition of DNA: An NMR Spectroscopic Study
Chemistry, 8, 2002
|
|
1I5W
 
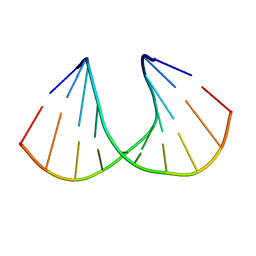 | | A-DNA DECAMER GCGTA(TLN)ACGC | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(TLN)P*AP*CP*GP*C)-3' | | Authors: | Egli, M, Minasov, G, Teplova, M, Kumar, R, Wengel, J. | | Deposit date: | 2001-03-01 | | Release date: | 2001-04-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray crystal structure of a locked nucleic acid (LNA) duplex composed of a palindromic 10-mer DNA strand containing one LNA thymine monomer
J.Chem.Soc.,Chem.Commun., 2001
|
|
1KLA
 
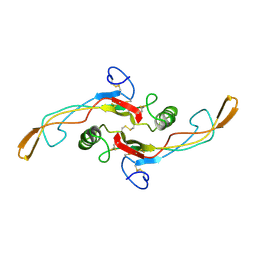 | | SOLUTION STRUCTURE OF TGF-B1, NMR, MODELS 1-17 OF 33 STRUCTURES | | Descriptor: | TRANSFORMING GROWTH FACTOR-BETA 1 | | Authors: | Hinck, A.P, Archer, S.J, Qian, S.W, Roberts, A.B, Sporn, M.B, Weatherbee, J.A, Tsang, M.L.-S, Lucas, R, Zhang, B.-L, Wenker, J, Torchia, D.A. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2.
Biochemistry, 35, 1996
|
|
1KLC
 
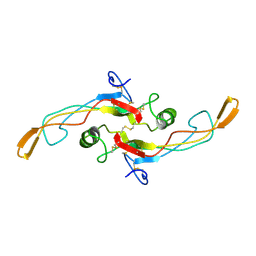 | | SOLUTION STRUCTURE OF TGF-B1, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TRANSFORMING GROWTH FACTOR-BETA 1 | | Authors: | Hinck, A.P, Archer, S.J, Qian, S.W, Roberts, A.B, Sporn, M.B, Weatherbee, J.A, Tsang, M.L.-S, Lucas, R, Zhang, B.-L, Wenker, J, Torchia, D.A. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2.
Biochemistry, 35, 1996
|
|
1KLD
 
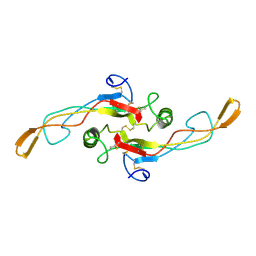 | | SOLUTION STRUCTURE OF TGF-B1, NMR, MODELS 18-33 OF 33 STRUCTURES | | Descriptor: | TRANSFORMING GROWTH FACTOR-BETA 1 | | Authors: | Hinck, A.P, Archer, S.J, Qian, S.W, Roberts, A.B, Sporn, M.B, Weatherbee, J.A, Tsang, M.L.-S, Lucas, R, Zhang, B.-L, Wenker, J, Torchia, D.A. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2.
Biochemistry, 35, 1996
|
|
3ENI
 
 | | Crystal structure of the Fenna-Matthews-Olson Protein from Chlorobaculum Tepidum | | Descriptor: | BACTERIOCHLOROPHYLL A, Bacteriochlorophyll a protein | | Authors: | Tronrud, D, Camara-Artigas, A, Blankenship, R, Allen, J.P. | | Deposit date: | 2008-09-25 | | Release date: | 2009-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structural basis for the difference in absorbance spectra for the FMO antenna protein from various green sulfur bacteria.
Photosynth.Res., 100, 2009
|
|
4KQA
 
 | | Crystal structure of the golgi casein kinase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NICKEL (II) ION, Protein H03A11.1, ... | | Authors: | Xiao, J. | | Deposit date: | 2013-05-14 | | Release date: | 2013-06-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Crystal structure of the Golgi casein kinase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KQB
 
 | | crystal structure of the Golgi casein kinase with Mn/ADP bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Xiao, J. | | Deposit date: | 2013-05-14 | | Release date: | 2013-06-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.045 Å) | | Cite: | Crystal structure of the Golgi casein kinase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8GQE
 
 | | Crystal structure of the W285A mutant of UVR8 in complex with RUP2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ultraviolet-B receptor UVR8, WD repeat-containing protein RUP2 | | Authors: | Wang, Y.D, Wang, L.X, Guan, Z.Y, chang, H.F, Yin, P. | | Deposit date: | 2022-08-30 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RUP2 facilitates UVR8 redimerization via two interfaces.
Plant Commun., 4, 2023
|
|
7XC2
 
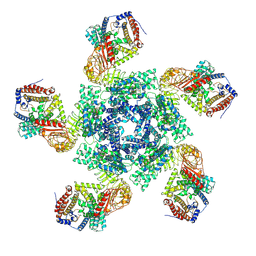 | | Cryo EM structure of oligomeric complex formed by wheat CNL Sr35 and the effector AvrSr35 of the wheat stem rust pathogen | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Avirulence factor, CNL9 | | Authors: | Alexander, F, Li, E.T, Aaron, L, Deng, Y.N, Sun, Y, Chai, J.J. | | Deposit date: | 2022-03-22 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A wheat resistosome defines common principles of immune receptor channels.
Nature, 610, 2022
|
|
7DP8
 
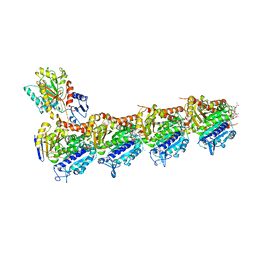 | | Crystal structure of T2R-TTL-Cevipabulin-eribulin complex | | Descriptor: | (1S,3S,6S,9S,12S,14R,16R,18S,20R,21R,22S,26R,29S,31R,32S,33R,35R,36S)-20-[(2S)-3-amino-2-hydroxypropyl]-21-methoxy-14-methyl-8,15-dimethylidene-2,19,30,34,37,39,40,41-octaoxanonacyclo[24.9.2.1~3,32~.1~3,33~.1~6,9~.1~12,16~.0~18,22~.0~29,36~.0~31,35~]hentetracontan-24-one (non-preferred name), 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[2,6-bis(fluoranyl)-4-[3-(methylamino)propoxy]phenyl]-5-chloranyl-N-[(2S)-1,1,1-tris(fluoranyl)propan-2-yl]-[1,2,4]triazolo[1,5-a]pyrimidin-7-amine, ... | | Authors: | Chen, L.J, Chen, Q, Yu, Y, Yang, J.H. | | Deposit date: | 2020-12-18 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.446 Å) | | Cite: | Cevipabulin-tubulin complex reveals a novel agent binding site on alpha-tubulin with tubulin degradation effect.
Sci Adv, 7, 2021
|
|
7CLD
 
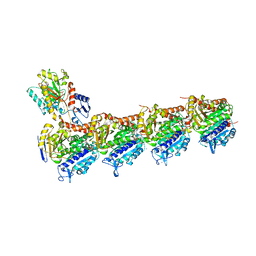 | | Crystal structure of T2R-TTL-Cevipabulin complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[2,6-bis(fluoranyl)-4-[3-(methylamino)propoxy]phenyl]-5-chloranyl-N-[(2S)-1,1,1-tris(fluoranyl)propan-2-yl]-[1,2,4]triazolo[1,5-a]pyrimidin-7-amine, CALCIUM ION, ... | | Authors: | Chen, L.J, Chen, Q, Yu, Y, Yang, J.H. | | Deposit date: | 2020-07-20 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.611 Å) | | Cite: | Cevipabulin-tubulin complex reveals a novel agent binding site on alpha-tubulin with tubulin degradation effect.
Sci Adv, 7, 2021
|
|
7CXZ
 
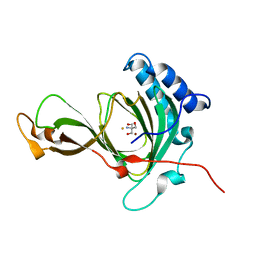 | | crystal structure of pco2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE (III) ION, NICKEL (II) ION, ... | | Authors: | Guo, Q, Xu, C, Liao, S, Chen, Z. | | Deposit date: | 2020-09-02 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | Molecular basis for cysteine oxidation by plant cysteine oxidases from Arabidopsis thaliana.
J.Struct.Biol., 213, 2021
|
|
7CHJ
 
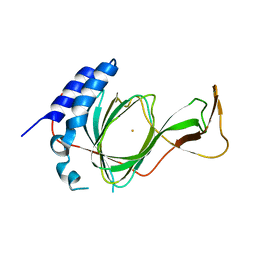 | | crystal structure of pco4 | | Descriptor: | CITRIC ACID, FE (III) ION, Plant cysteine oxidase 4 | | Authors: | Guo, Q, Xu, C, Liao, S. | | Deposit date: | 2020-07-05 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Molecular basis for cysteine oxidation by plant cysteine oxidases from Arabidopsis thaliana.
J.Struct.Biol., 213, 2021
|
|
7CHI
 
 | | crystal structure of pco5 | | Descriptor: | FE (III) ION, Plant cysteine oxidase 5 | | Authors: | Guo, Q, Xu, C, Liao, S. | | Deposit date: | 2020-07-05 | | Release date: | 2020-11-25 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Molecular basis for cysteine oxidation by plant cysteine oxidases from Arabidopsis thaliana.
J.Struct.Biol., 213, 2021
|
|
4LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
5FXR
 
 | | IGFR-1R complex with a pyrimidine inhibitor. | | Descriptor: | 5-chloranyl-4-imidazo[1,2-a]pyridin-3-yl-N-(3-methyl-1-piperidin-4-yl-pyrazol-4-yl)pyrimidin-2-amine, INSULIN-LIKE GROWTH FACTOR 1 RECEPTOR | | Authors: | Degorce, S, Boyd, S, Curwen, J, Ducray, R, Halsall, C, Jones, C, Lach, F, Lenz, E, Pass, M, Pass, S, Trigwell, C, Norman, R, Phillips, C. | | Deposit date: | 2016-03-02 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Potent, Selective, Orally Bioavailable, and Efficacious Novel 2-(Pyrazol-4-ylamino)-pyrimidine Inhibitor of the Insulin-like Growth Factor-1 Receptor (IGF-1R).
J. Med. Chem., 59, 2016
|
|
5FXQ
 
 | | IGFR-1R complex with a pyrimidine inhibitor. | | Descriptor: | 5-chloranyl-4-imidazo[1,2-a]pyridin-3-yl-N-(5-methyl-1-piperidin-4-yl-pyrazol-4-yl)pyrimidin-2-amine, INSULIN-LIKE GROWTH FACTOR 1 RECEPTOR | | Authors: | Degorce, S, Boyd, S, Curwen, J, Ducray, R, Halsall, C, Jones, C, Lach, F, Lenz, E, Pass, M, Pass, S, Trigwell, C, Norman, R, Phillips, C. | | Deposit date: | 2016-03-02 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a Potent, Selective, Orally Bioavailable, and Efficacious Novel 2-(Pyrazol-4-ylamino)-pyrimidine Inhibitor of the Insulin-like Growth Factor-1 Receptor (IGF-1R).
J. Med. Chem., 59, 2016
|
|
5FXS
 
 | | IGFR-1R complex with a pyrimidine inhibitor. | | Descriptor: | 2-[4-[4-[[(6Z)-5-chloranyl-6-pyrazolo[1,5-a]pyridin-3-ylidene-1H-pyrimidin-2-yl]amino]-3,5-dimethyl-pyrazol-1-yl]piperidin-1-yl]-N,N-dimethyl-ethanamide, ACETATE ION, INSULIN-LIKE GROWTH FACTOR 1 RECEPTOR | | Authors: | Degorce, S, Boyd, S, Curwen, J, Ducray, R, Halsall, C, Jones, C, Lach, F, Lenz, E, Pass, M, Pass, S, Trigwell, C, Norman, R, Phillips, C. | | Deposit date: | 2016-03-02 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a Potent, Selective, Orally Bioavailable, and Efficacious Novel 2-(Pyrazol-4-ylamino)-pyrimidine Inhibitor of the Insulin-like Growth Factor-1 Receptor (IGF-1R).
J. Med. Chem., 59, 2016
|
|
5IW4
 
 | |
5IW5
 
 | |
3U4E
 
 | | Crystal Structure of PG9 Fab in Complex with V1V2 Region from HIV-1 strain CAP45 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PG9 Heavy Chain, PG9 Light Chain, ... | | Authors: | Gorman, J, McLellan, J, Pancera, M, Kwong, P.D. | | Deposit date: | 2011-10-07 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.185 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
