7MBJ
 
 | |
5YBC
 
 | | X-ray structure of native ETS-domain domain of Ergp55 | | Descriptor: | GLYCEROL, Transcriptional regulator ERG | | Authors: | Saxena, A.K, Gangwar, S.P. | | Deposit date: | 2017-09-04 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Comparative structure analysis of the ETSi domain of ERG3 and its complex with the E74 promoter DNA sequence
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5YBD
 
 | | X-ray structure of ETS domain of Ergp55 in complex with E74DNA | | Descriptor: | DNA (5'-D(P*AP*CP*CP*GP*GP*AP*AP*GP*T)-3'), DNA (5'-D(P*CP*AP*CP*TP*TP*CP*CP*GP*GP*T)-3'), Transcriptional regulator ERG | | Authors: | Saxena, A.K, Gangwar, S.P. | | Deposit date: | 2017-09-04 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.769 Å) | | Cite: | Comparative structure analysis of the ETSi domain of ERG3 and its complex with the E74 promoter DNA sequence
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6XMX
 
 | | Cryo-EM structure of BCL6 bound to BI-3802 | | Descriptor: | 2-[6-[[5-chloranyl-2-[(3~{S},5~{R})-3,5-dimethylpiperidin-1-yl]pyrimidin-4-yl]amino]-1-methyl-2-oxidanylidene-quinolin-3-yl]oxy-~{N}-methyl-ethanamide, B-cell lymphoma 6 protein | | Authors: | Yoon, H, Burman, S.S.R, Hunkeler, M, Nowak, R.P, Fischer, E.S. | | Deposit date: | 2020-07-01 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Small-molecule-induced polymerization triggers degradation of BCL6.
Nature, 588, 2020
|
|
6X8E
 
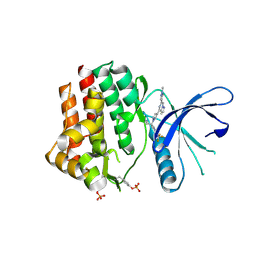 | | Crystal structure of JAK2 with Compound 11 | | Descriptor: | Tyrosine-protein kinase JAK2, [3-{4-[6-(1-methyl-1H-pyrazol-4-yl)pyrazolo[1,5-a]pyrazin-4-yl]-1H-pyrazol-1-yl}-1-(2,2,2-trifluoroethyl)azetidin-3-yl]acetonitrile | | Authors: | Vajdos, F.F, Knafels, J.D. | | Deposit date: | 2020-06-01 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Tyrosine Kinase 2 (TYK2) Inhibitor (PF-06826647) for the Treatment of Autoimmune Diseases.
J.Med.Chem., 63, 2020
|
|
6X8G
 
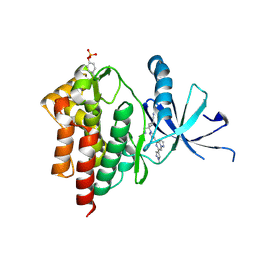 | | Crystal structure of TYK2 with Compound 22 | | Descriptor: | Non-receptor tyrosine-protein kinase TYK2, trans-3-(cyanomethyl)-3-{4-[6-(1-methyl-1H-pyrazol-4-yl)pyrazolo[1,5-a]pyrazin-4-yl]-1H-pyrazol-1-yl}cyclobutane-1-carbonitrile | | Authors: | Vajdos, F.F, Knafels, J.D. | | Deposit date: | 2020-06-01 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery of Tyrosine Kinase 2 (TYK2) Inhibitor (PF-06826647) for the Treatment of Autoimmune Diseases.
J.Med.Chem., 63, 2020
|
|
6X8F
 
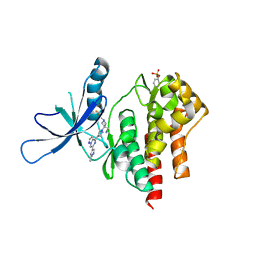 | | Crystal structure of TYK2 with Compound 11 | | Descriptor: | Non-receptor tyrosine-protein kinase TYK2, [3-{4-[6-(1-methyl-1H-pyrazol-4-yl)pyrazolo[1,5-a]pyrazin-4-yl]-1H-pyrazol-1-yl}-1-(2,2,2-trifluoroethyl)azetidin-3-yl]acetonitrile | | Authors: | Vajdos, F.F, Knafels, J.D. | | Deposit date: | 2020-06-01 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of Tyrosine Kinase 2 (TYK2) Inhibitor (PF-06826647) for the Treatment of Autoimmune Diseases.
J.Med.Chem., 63, 2020
|
|
4N7W
 
 | | Crystal Structure of the sodium bile acid symporter from Yersinia frederiksenii | | Descriptor: | CITRIC ACID, Transporter, sodium/bile acid symporter family, ... | | Authors: | Zhou, X, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2013-10-16 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural basis of the alternating-access mechanism in a bile acid transporter.
Nature, 505, 2013
|
|
8I60
 
 | |
3ZTZ
 
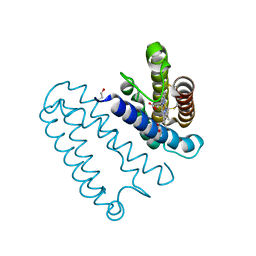 | | Cytochrome c prime from alcaligenes xylosoxidans: carbon monooxide bound L16G variant at 1.05 A resolution: unrestraint refinement | | Descriptor: | CARBON MONOXIDE, CYTOCHROME C', HEME C | | Authors: | Antonyuk, S.V, Rustage, N, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2011-07-12 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Carbon Monoxide Poisoning is Prevented by the Energy Costs of Conformational Changes in Gas- Binding Haemproteins.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3ZQV
 
 | |
3ZQY
 
 | | CYTOCHROME C PRIME FROM ALCALIGENES XYLOSOXIDANS: CARBON MONOOXIDE BOUND L16A VARIANT AT 1.03 A RESOLUTION- NON-RESTRAINT REFINEMENT | | Descriptor: | CARBON MONOXIDE, CYTOCHROME C', HEME C | | Authors: | Antonyuk, S.V, Rustage, N, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2011-06-12 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Carbon Monoxide Poisoning is Prevented by the Energy Costs of Conformational Changes in Gas- Binding Haemproteins.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3ZTM
 
 | | Cytochrome c prime from alcaligenes xylosoxidans: as isolated L16G variant at 0.9 A resolution: unrestraint refinement | | Descriptor: | CARBON MONOXIDE, CYTOCHROME C', HEME C | | Authors: | Antonyuk, S.V, Rustage, N, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2011-07-11 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Carbon Monoxide Poisoning is Prevented by the Energy Costs of Conformational Changes in Gas- Binding Haemproteins.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3ZWI
 
 | | RECOMBINANT NATIVE CYTOCHROME C PRIME FROM ALCALIGENES XYLOSOXIDANS: CARBON MONOOXIDE BOUND AT 1.25 A:UNRESTRAINT REFINEMENT | | Descriptor: | ASCORBIC ACID, CARBON MONOXIDE, CYTOCHROME C', ... | | Authors: | Antonyuk, S, Rustage, N, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2011-07-31 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Carbon Monoxide Poisoning is Prevented by the Energy Costs of Conformational Changes in Gas-Binding Haemproteins.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7PQE
 
 | |
7JY1
 
 | | Structure of HbA with compound 19 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Jasti, J. | | Deposit date: | 2020-08-28 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | PF-07059013: A Noncovalent Modulator of Hemoglobin for Treatment of Sickle Cell Disease.
J.Med.Chem., 64, 2021
|
|
5YC9
 
 | |
7JY0
 
 | | Structure of HbA with compound 9 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 2-amino-3-{(1S)-1-[5-fluoro-2-(1H-pyrazol-1-yl)phenyl]ethoxy}quinoline-6-carboxamide, CARBON MONOXIDE, ... | | Authors: | Jasti, J. | | Deposit date: | 2020-08-28 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | PF-07059013: A Noncovalent Modulator of Hemoglobin for Treatment of Sickle Cell Disease.
J.Med.Chem., 64, 2021
|
|
7JXZ
 
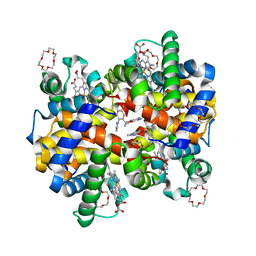 | | Structure of HbA with compound (S)-4 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 3-{(1S)-1-[5-fluoro-2-(1H-pyrazol-1-yl)phenyl]ethoxy}-5-(3-methyl-1H-pyrazol-4-yl)pyridin-2-amine, CARBON MONOXIDE, ... | | Authors: | Jasti, J. | | Deposit date: | 2020-08-28 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | PF-07059013: A Noncovalent Modulator of Hemoglobin for Treatment of Sickle Cell Disease.
J.Med.Chem., 64, 2021
|
|
7PPO
 
 | |
5ULA
 
 | |
4N7X
 
 | |
4Z0M
 
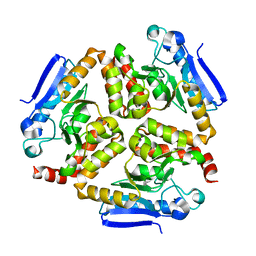 | | EchA5 Mycobacterium tuberculosis | | Descriptor: | Enoyl-CoA hydratase | | Authors: | Chaudhary, S, Gokhale, R.S. | | Deposit date: | 2015-03-26 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Unsaturated Lipid Assimilation by Mycobacteria Requires Auxiliary cis-trans Enoyl CoA Isomerase
Chem.Biol., 22, 2015
|
|
5E0N
 
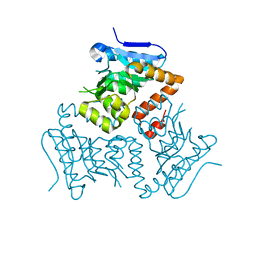 | | Crystal Structure of MSMEG_3139, a monofunctional enoyl CoA isomerase from M.smegmatis | | Descriptor: | Enoyl-CoA hydratase/isomerase | | Authors: | Priyadarshan, K, Haque, A.S, Anandakrishnan, M, Sankaranarayanan, R. | | Deposit date: | 2015-09-29 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.061 Å) | | Cite: | Unsaturated Lipid Assimilation by Mycobacteria Requires Auxiliary cis-trans Enoyl CoA Isomerase.
Chem.Biol., 22, 2015
|
|
6QIY
 
 | | CI-2, conformation 1 | | Descriptor: | Subtilisin-chymotrypsin inhibitor-2A | | Authors: | Romero, A, Ruiz, F.M. | | Deposit date: | 2019-01-21 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering protein assemblies with allosteric control via monomer fold-switching.
Nat Commun, 10, 2019
|
|
