8DYD
 
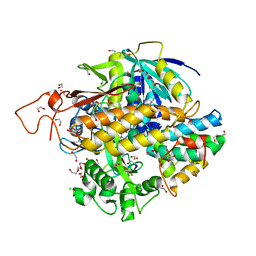 | | Crystal structure of human SDHA-SDHAF2-SDHAF4 assembly intermediate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sharma, P, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2022-08-04 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Disordered-to-ordered transitions in assembly factors allow the complex II catalytic subunit to switch binding partners.
Nat Commun, 15, 2024
|
|
8DYE
 
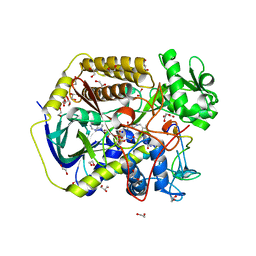 | | Crystal structure of human SDHA-SDHAF4 assembly intermediate | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Sharma, P, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2022-08-04 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Disordered-to-ordered transitions in assembly factors allow the complex II catalytic subunit to switch binding partners.
Nat Commun, 15, 2024
|
|
3SP3
 
 | | Lysozyme in 20% sucrose | | Descriptor: | CHLORIDE ION, Lysozyme, SODIUM ION, ... | | Authors: | Sharma, P, Solanki, A.K, Ashish | | Deposit date: | 2011-07-01 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Lysozyme in 20% sucrose
to be published
|
|
4D9Z
 
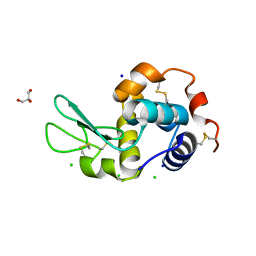 | | Lysozyme at 318K | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Sharma, P, Ashish | | Deposit date: | 2012-01-12 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.709 Å) | | Cite: | Characterization of heat induced spherulites of lysozyme reveals new insight on amyloid initiation
Sci Rep, 6, 2016
|
|
4DC4
 
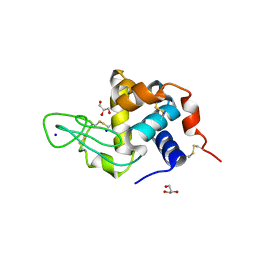 | | Lysozyme Trimer | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Sharma, P, Ashish | | Deposit date: | 2012-01-17 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.654 Å) | | Cite: | Characterization of heat induced spherulites of lysozyme reveals new insight on amyloid initiation
Sci Rep, 6, 2016
|
|
4EOF
 
 | | Lysozyme in the presence of arginine | | Descriptor: | ACETATE ION, ARGININE, CHLORIDE ION, ... | | Authors: | Sharma, P, Ashish | | Deposit date: | 2012-04-14 | | Release date: | 2013-04-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Characterization of heat induced spherulites of lysozyme reveals new insight on amyloid initiation.
Sci Rep, 6, 2016
|
|
5CBP
 
 | | Crystal Structure of Conjoint Pyrococcus furiosus L-asparaginase at 37 degree C | | Descriptor: | CITRATE ANION, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Sharma, P, Yadav, S.P, Tomar, R, Kundu, B, Ashish, F. | | Deposit date: | 2015-07-01 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.358 Å) | | Cite: | Heat induces end to end repetitive association in P. furiosus L-asparaginase which enables its thermophilic property.
Sci Rep, 10, 2020
|
|
4HSF
 
 | | Lysozyme with Arginine at 318K | | Descriptor: | ARGININE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sharma, P, Ashish, F. | | Deposit date: | 2012-10-30 | | Release date: | 2013-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Lysozyme with Arginine at 318K.
To be Published
|
|
6B58
 
 | | FrdA-SdhE assembly intermediate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sharma, P, Iverson, T.M. | | Deposit date: | 2017-09-28 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.611 Å) | | Cite: | Crystal structure of an assembly intermediate of respiratory Complex II.
Nat Commun, 9, 2018
|
|
4II8
 
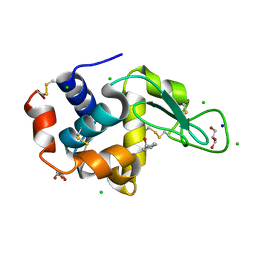 | | Lysozyme with Benzyl alcohol | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Sharma, P, Ashish | | Deposit date: | 2012-12-20 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Characterization of heat induced spherulites of lysozyme reveals new insight on amyloid initiation
Sci Rep, 6, 2016
|
|
3RT5
 
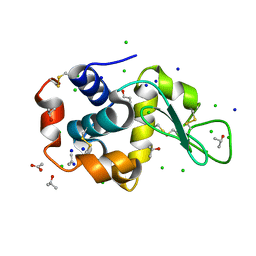 | | Lysozyme in 30% propanol | | Descriptor: | ACETATE ION, CHLORIDE ION, ISOPROPYL ALCOHOL, ... | | Authors: | Sharma, P, Solanki, A.K, Ashish | | Deposit date: | 2011-05-03 | | Release date: | 2011-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Lysozyme in 30% propanol
to be published
|
|
3RNX
 
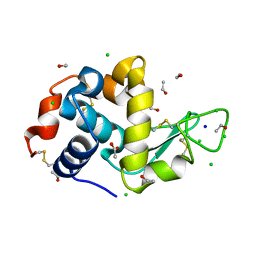 | |
3RW8
 
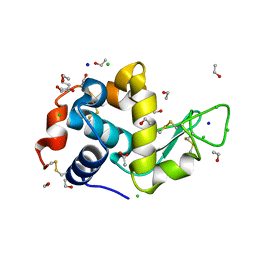 | |
3UIL
 
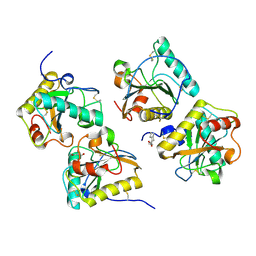 | | Crystal Structure of the complex of PGRP-S with lauric acid at 2.2 A resolution | | Descriptor: | GLYCEROL, LAURIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-11-05 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site
Arch.Biochem.Biophys., 529, 2013
|
|
3UMQ
 
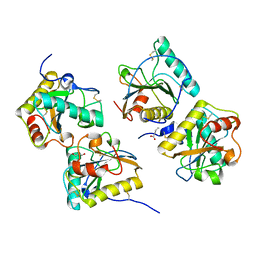 | | Crystal structure of peptidoglycan recognition protein-S complexed with butyric acid at 2.2 A resolution | | Descriptor: | GLYCEROL, Peptidoglycan recognition protein 1, butanoic acid | | Authors: | Pandey, N, Sharma, P, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-11-14 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site
Arch.Biochem.Biophys., 529, 2013
|
|
3USX
 
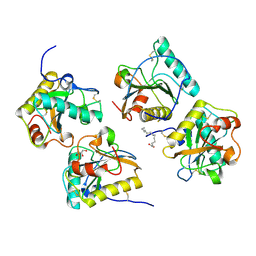 | | Crystal structure of PGRP-S complexed with Myristic Acid at 2.28 A resolution | | Descriptor: | GLYCEROL, MYRISTIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Yamini, S, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-11-24 | | Release date: | 2012-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site
Arch.Biochem.Biophys., 529, 2013
|
|
4FNN
 
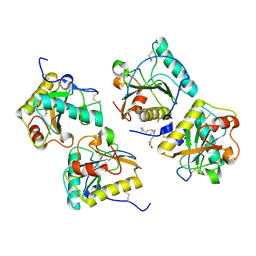 | | Crystal structure of the complex of CPGRP-S with stearic acid at 2.2 A RESOLUTION | | Descriptor: | Peptidoglycan recognition protein 1, STEARIC ACID | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-06-20 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site.
Arch.Biochem.Biophys., 529, 2013
|
|
6UH8
 
 | | Crystal structure of DAD2 N242I mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Decreased Apical Dominance 2, GLYCEROL, ... | | Authors: | Sharma, P, Hamiaux, C, Snowden, K.C. | | Deposit date: | 2019-09-27 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Flexibility of the petunia strigolactone receptor DAD2 promotes its interaction with signaling partners.
J.Biol.Chem., 295, 2020
|
|
6UH9
 
 | | Crystal structure of DAD2 D166A mutant | | Descriptor: | Decreased Apical Dominance 2, TETRAETHYLENE GLYCOL | | Authors: | Sharma, P, Hamiaux, C, Snowden, K.C. | | Deposit date: | 2019-09-27 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Flexibility of the petunia strigolactone receptor DAD2 promotes its interaction with signaling partners.
J.Biol.Chem., 295, 2020
|
|
5GVY
 
 | | Crystal structure of SALT protein from Oryza sativa | | Descriptor: | Salt stress-induced protein, alpha-D-mannopyranose | | Authors: | Sharma, P, Sagar, A, Kaur, N, Sharma, I, Kirat, K, Ashish, F.N.U, Pati, P.K. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.662 Å) | | Cite: | Structural insights into rice SalTol QTL located SALT protein.
Sci Rep, 10, 2020
|
|
4RA6
 
 | | Crystal structure of linker less Pyrococcus furiosus L-asparaginase | | Descriptor: | GLYCEROL, L-asparaginase | | Authors: | Sharma, P, Tomar, R, Ashish, Kundu, B. | | Deposit date: | 2014-09-09 | | Release date: | 2014-12-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Structural and functional insights into an archaeal L-asparaginase obtained through the linker-less assembly of constituent domains.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8SHH
 
 | | Crystal structure of EvdS6 decarboxylase in ligand free state | | Descriptor: | DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, dTDP-glucose 4,6-dehydratase | | Authors: | Sharma, P, Frigo, L, Dulin, C.C, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2023-04-14 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | EvdS6 is a bifunctional decarboxylase from the everninomicin gene cluster.
J.Biol.Chem., 299, 2023
|
|
8SK0
 
 | | Crystal structure of EvdS6 decarboxylase in ligand bound state | | Descriptor: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sharma, P, Frigo, L, Dulin, C.C, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2023-04-18 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | EvdS6 is a bifunctional decarboxylase from the everninomicin gene cluster.
J.Biol.Chem., 299, 2023
|
|
4NJE
 
 | | Crystal structure of Pyrococcus furiosus L-asparaginase with ligand | | Descriptor: | ASPARTIC ACID, L-asparaginase | | Authors: | Sharma, P, Tomar, R, Singh, S, Yadav, S.P.S, Ashish, Kundu, B. | | Deposit date: | 2013-11-09 | | Release date: | 2014-12-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional insights into an archaeal L-asparaginase obtained through the linker-less assembly of constituent domains.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q0M
 
 | | Crystal structure of Pyrococcus furiosus L-asparaginase | | Descriptor: | (4S)-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, GLYCEROL, L-asparaginase, ... | | Authors: | Sharma, P, Tomar, R, Singh, S, Yadav, S.P.S, Ashish, Kundu, B. | | Deposit date: | 2014-04-02 | | Release date: | 2014-12-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.226 Å) | | Cite: | Structural and functional insights into an archaeal L-asparaginase obtained through the linker-less assembly of constituent domains.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
