2ZOW
 
 | | Crystal Structure of H2O2 treated Cu,Zn-SOD | | Descriptor: | COPPER (I) ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Ito, S, Ishii, T, Sakai, H, Uchida, K. | | Deposit date: | 2008-06-11 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structures of H2O2-treated Cu,Zn-superoxide dismutase
To be Published
|
|
2ZBR
 
 | | Crystal structure of ribosomal protein L11 methyltransferase from Thermus thermophilus in complex with S-adenosyl-ornithine | | Descriptor: | Ribosomal protein L11 methyltransferase, SINEFUNGIN | | Authors: | Kaminishi, T, Sakai, H, Takemoto-Hori, C, Terada, T, Nakagawa, N, Maoka, N, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-10-26 | | Release date: | 2008-11-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of ribosomal protein L11 methyltransferase from Thermus thermophilus
To be Published
|
|
2ZC7
 
 | |
1IUH
 
 | | Crystal structure of TT0787 of thermus thermophilus HB8 | | Descriptor: | 2'-5' RNA Ligase | | Authors: | Kato, M, Sakai, H, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-05 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the 2'-5' RNA Ligase from Thermus thermophilus HB8
J.MOL.BIOL., 329, 2003
|
|
1UKL
 
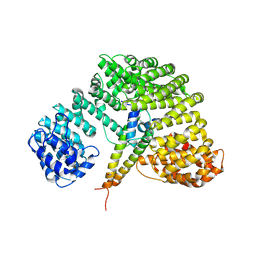 | | Crystal structure of Importin-beta and SREBP-2 complex | | Descriptor: | Importin beta-1 subunit, Sterol regulatory element binding protein-2 | | Authors: | Lee, S.J, Sekimoto, T, Yamashita, E, Nagoshi, E, Nakagawa, A, Imamoto, N, Yoshimura, M, Sakai, H, Tsukihara, T, Yoneda, Y. | | Deposit date: | 2003-08-26 | | Release date: | 2003-12-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of Importin-beta Bound to SREBP-2: Nuclear Import of a Transcription Factor
Science, 302, 2003
|
|
1WD7
 
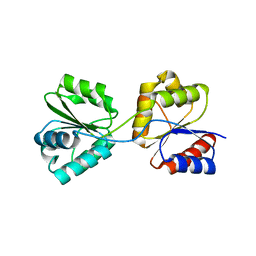 | | Crystal Structure of Uroporphyrinogen III Synthase from an Extremely Thermophilic Bacterium Thermus thermophilus HB8 (Wild type, Native, Form-2 crystal) | | Descriptor: | Uroporphyrinogen III Synthase | | Authors: | Mizohata, E, Matsuura, T, Sakai, H, Murayama, K, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-12 | | Release date: | 2004-11-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Uroporphyrinogen III Synthase from an Extremely Thermophilic Bacterium Thermus thermophilus HB8 (Wild type, Native, Form-2 crystal)
to be published
|
|
1WKY
 
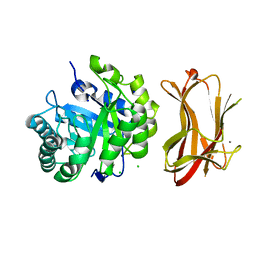 | | Crystal structure of alkaline mannanase from Bacillus sp. strain JAMB-602: catalytic domain and its Carbohydrate Binding Module | | Descriptor: | CALCIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Akita, M, Takeda, N, Hirasawa, K, Sakai, H, Kawamoto, M, Yamamoto, M, Grant, W.D, Hatada, Y, Ito, S, Horikoshi, K. | | Deposit date: | 2004-06-15 | | Release date: | 2005-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallization and preliminary X-ray study of alkaline mannanase from an alkaliphilic Bacillus isolate.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1WCX
 
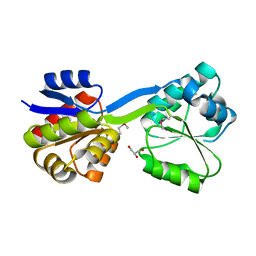 | | Crystal Structure of Mutant Uroporphyrinogen III Synthase from an Extremely Thermophilic Bacterium Thermus thermophilus HB8 (L75M/I193M/L248M, SeMet derivative, Form-1 crystal) | | Descriptor: | GLYCEROL, Uroporphyrinogen III Synthase | | Authors: | Mizohata, E, Matsuura, T, Murayama, K, Sakai, H, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-06 | | Release date: | 2005-05-06 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Uroporphyrinogen III Synthase from Thermus thermophilus HB8
To be Published
|
|
1WCW
 
 | | Crystal Structure of Uroporphyrinogen III Synthase from an Extremely Thermophilic Bacterium Thermus thermophilus HB8 (Wild type, Native, Form-1 crystal) | | Descriptor: | GLYCEROL, Uroporphyrinogen III synthase | | Authors: | Mizohata, E, Matsuura, T, Sakai, H, Murayama, K, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-06 | | Release date: | 2005-05-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Uroporphyrinogen III Synthase from Thermus thermophilus HB8
To be Published
|
|
1WK4
 
 | | Crystal structure of ttk003001606 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ttk003001606 | | Authors: | Kaminishi, T, Sakai, H, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-30 | | Release date: | 2004-11-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ttk003001606
To be Published
|
|
1WWM
 
 | | Crystal Structure of Conserved Hypothetical Protein TT2028 from an Extremely Thermophilic Bacterium Thermus thermophilus HB8 | | Descriptor: | hypothetical protein TT2028 | | Authors: | Mizohata, E, Ushikoshi-Nakayama, R, Terada, T, Murayama, K, Sakai, H, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-07 | | Release date: | 2005-07-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structure of TT2028 from an Extremely Thermophilic Bacterium Thermus thermophilus HB8
To be Published
|
|
1YQZ
 
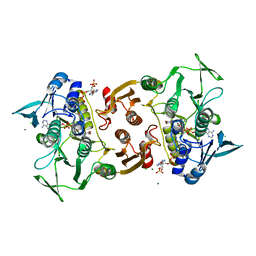 | | Structure of Coenzyme A-Disulfide Reductase from Staphylococcus aureus refined at 1.54 Angstrom resolution | | Descriptor: | CHLORIDE ION, COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mallett, T.C, Wallen, J.R, Sakai, H, Luba, J, Parsonage, D, Karplus, P.A, Tsukihara, T, Claiborne, A. | | Deposit date: | 2005-02-02 | | Release date: | 2006-05-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure of coenzyme A-disulfide reductase from Staphylococcus aureus at 1.54 A resolution.
Biochemistry, 45, 2006
|
|
1GCJ
 
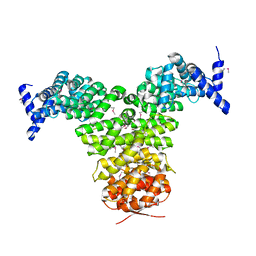 | | N-TERMINAL FRAGMENT OF IMPORTIN-BETA | | Descriptor: | IMPORTIN BETA | | Authors: | Lee, S.J, Imamoto, N, Sakai, H, Nakagawa, A, Kose, S, Koike, M, Yamamoto, M, Kumasaka, T, Yoneda, Y, Tsukihara, T. | | Deposit date: | 2000-07-31 | | Release date: | 2000-10-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The adoption of a twisted structure of importin-beta is essential for the protein-protein interaction required for nuclear transport.
J.Mol.Biol., 302, 2000
|
|
1IQQ
 
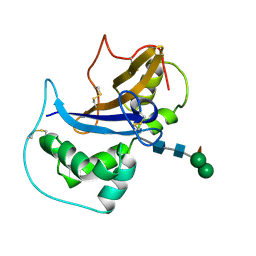 | | Crystal Structure of Japanese pear S3-RNase | | Descriptor: | S3-RNase, beta-D-xylopyranose-(1-2)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Matsuura, T, Sakai, H, Norioka, S. | | Deposit date: | 2001-07-25 | | Release date: | 2001-11-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure at 1.5-A resolution of Pyrus pyrifolia pistil ribonuclease responsible for gametophytic self-incompatibility.
J.Biol.Chem., 276, 2001
|
|
1J3U
 
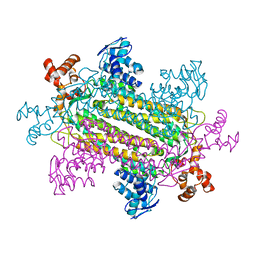 | |
1K9A
 
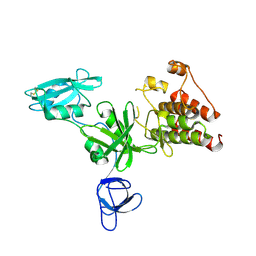 | | Crystal structure analysis of full-length carboxyl-terminal Src kinase at 2.5 A resolution | | Descriptor: | Carboxyl-terminal Src kinase | | Authors: | Ogawa, A, Takayama, Y, Nagata, A, Chong, K.T, Takeuchi, S, Sakai, H, Nakagawa, A, Nada, S, Okada, M, Tsukihara, T. | | Deposit date: | 2001-10-28 | | Release date: | 2002-03-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the carboxyl-terminal Src kinase, Csk.
J.Biol.Chem., 277, 2002
|
|
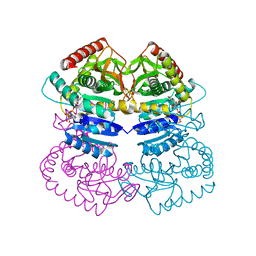
1LLD
 
 | |
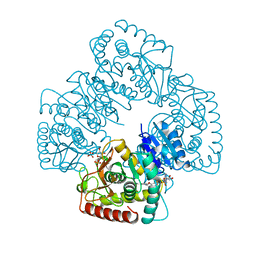
1LTH
 
 | |
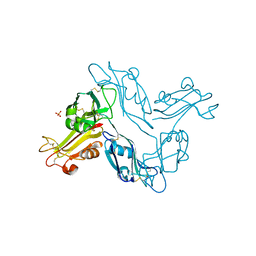
5H4S
 
 | |
6AHS
 
 | | Mouse Kallikrein 7 in complex with imidazolinylindole derivative | | Descriptor: | 1-[(2-chlorophenyl)sulfonyl]-5-methyl-3-[(4R)-2-methyl-4,5-dihydro-1H-imidazol-4-yl]-1H-indole, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Sugawara, H. | | Deposit date: | 2018-08-20 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery and structure-activity relationship of imidazolinylindole derivatives as kallikrein 7 inhibitors.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
5ZFI
 
 | | Mouse kallikrein 7 in complex with 6-benzyl-1,4-diazepan-7-one derivative | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[(3Z,6R)-6-[(2,6-dichlorophenyl)methyl]-3-(dimethylhydrazinylidene)-7-oxo-1,4-diazepan-1-yl]-N-[3-(1-methyl-1H-pyrazol-4-yl)phenyl]acetamide, Kallikrein-7 | | Authors: | Sugawara, H. | | Deposit date: | 2018-03-06 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based drug design to overcome species differences in kallikrein 7 inhibition of 1,3,6-trisubstituted 1,4-diazepan-7-ones.
Bioorg. Med. Chem., 26, 2018
|
|
5ZFH
 
 | | Mouse Kallikrein 7 | | Descriptor: | Kallikrein-7 | | Authors: | Sugawara, H. | | Deposit date: | 2018-03-06 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure-based drug design to overcome species differences in kallikrein 7 inhibition of 1,3,6-trisubstituted 1,4-diazepan-7-ones.
Bioorg. Med. Chem., 26, 2018
|
|
1V8Q
 
 | | Crystal structure of ribosomal protein L27 from Thermus thermophilus HB8 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, TT0826 | | Authors: | Wang, H, Takemoto-Hori, C, Murayama, K, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-13 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ribosomal protein L27 from Thermus thermophilus HB8
Protein Sci., 13, 2004
|
|
2DDQ
 
 | |
5Y9L
 
 | | Human kallikrein 7 in complex with 1,3,6-trisubstituted 1,4-diazepane-7-one | | Descriptor: | 3-[2-[(3Z,6R)-6-[(5-chloranyl-2-methoxy-phenyl)methyl]-3-(dimethylhydrazinylidene)-7-oxidanylidene-1,4-diazepan-1-yl]ethanoylamino]benzoic acid, CHLORIDE ION, Kallikrein-7 | | Authors: | Sugawara, H. | | Deposit date: | 2017-08-25 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery and structure-activity relationship study of 1,3,6-trisubstituted 1,4-diazepane-7-ones as novel human kallikrein 7 inhibitors
Bioorg. Med. Chem. Lett., 27, 2017
|
|
