1PSY
 
 | | STRUCTURE OF RicC3, NMR, 20 STRUCTURES | | Descriptor: | 2S albumin | | Authors: | Pantoja-Uceda, D, Bruix, M, Gimenez-Gallego, G, Rico, M, Santoro, J. | | Deposit date: | 2003-06-22 | | Release date: | 2004-01-13 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RicC3, a 2S albumin storage protein from Ricinus communis.
Biochemistry, 42, 2003
|
|
2K11
 
 | | Solution structure of human pancreatic ribonuclease | | Descriptor: | Pancreatic Ribonuclease | | Authors: | Kover, K.E, Bruix, M, Santoro, J, Batta, G, Laurents, D.V, Rico, M. | | Deposit date: | 2008-02-20 | | Release date: | 2008-06-03 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure and dynamics of human pancreatic ribonuclease determined by NMR spectroscopy provide insight into its remarkable biological activities and inhibition.
J.Mol.Biol., 379, 2008
|
|
1S6D
 
 | | Structure in solution of a methionine-rich 2S Albumin protein from Sunflower Seed | | Descriptor: | Albumin 8 | | Authors: | Pantoja-Uceda, D, Bruix, M, Shewry, P.R, Santoro, J, Rico, M. | | Deposit date: | 2004-01-23 | | Release date: | 2004-06-29 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Methionine-Rich 2S Albumin from Sunflower Seeds: Relationship to Its Allergenic and Emulsifying Properties.
Biochemistry, 43, 2004
|
|
2JW4
 
 | | NMR solution structure of the N-terminal SH3 domain of human Nckalpha | | Descriptor: | Cytoplasmic protein NCK1 | | Authors: | Santiveri, C.M, Borroto, A, Simon, L, Rico, M, Ortiz, A.R, Alarcon, B, Jimenez, M. | | Deposit date: | 2007-10-05 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Interaction between the N-terminal SH3 domain of Nckalpha and CD3epsilon-derived peptides: Non-canonical and canonical recognition motifs
BIOCHEM.BIOPHYS.ACTA PROTEINS & PROTEOMICS, 1794, 2009
|
|
2JOI
 
 | | NMR solution structure of hypothetical protein TA0095 from Thermoplasma acidophilum | | Descriptor: | Hypothetical protein Ta0095 | | Authors: | Jimenez, M, Leon, E, Santoro, J, Rico, M, Yee, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-13 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the hypothetical protein TA0095 from Thermoplasma acidophilum: A novel superfamily with a two-layer sandwich architecture
Protein Sci., 16, 2007
|
|
1GL8
 
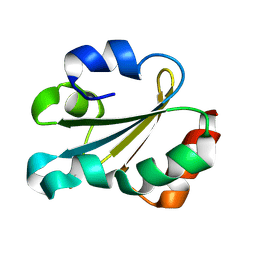 | | Solution structure of thioredoxin m from spinach, oxidized form | | Descriptor: | THIOREDOXIN | | Authors: | Neira, J.L, Gonzalez, C, Toiron, C, De-Prat-gay, G, Rico, M. | | Deposit date: | 2001-08-30 | | Release date: | 2001-12-13 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Solution Structure and Stability of Thioredoxin M from Spinach.
Biochemistry, 40, 2001
|
|
1H20
 
 | | Solution structure of the potato carboxypeptidase inhibitor | | Descriptor: | METALLOCARBOXYPEPTIDASE INHIBITOR | | Authors: | Gonzalez, C, Neira, J.L, Ventura, S, Bronsoms, S, Aviles, F.X, Rico, M. | | Deposit date: | 2002-07-29 | | Release date: | 2003-05-09 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of the Potato Carboxypeptidase Inhibitor by 1H and 15N NMR.
Proteins, 50, 2003
|
|
1PU1
 
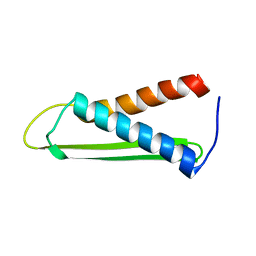 | | Solution structure of the Hypothetical protein mth677 from Methanothermobacter Thermautotrophicus | | Descriptor: | Hypothetical protein MTH677 | | Authors: | Blanco, F.J, Yee, A, Campos-Olivas, R, Devos, D, Valencia, A, Arrowsmith, C.H, Rico, M. | | Deposit date: | 2003-06-23 | | Release date: | 2004-06-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the hypothetical protein Mth677 from Methanobacterium thermoautotrophicum: A novel {alpha}+{beta} fold
Protein Sci., 13, 2004
|
|
4CCA
 
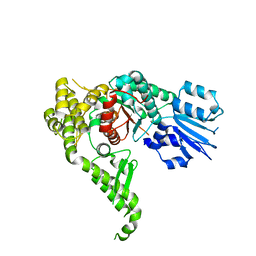 | | Structure of human Munc18-2 | | Descriptor: | CHLORIDE ION, SYNTAXIN-BINDING PROTEIN 2 | | Authors: | Hackmann, Y, Graham, S.C, Ehl, S, Hoening, S, Lehmberg, K, Arico, M, Owen, D.J, Griffiths, G.G. | | Deposit date: | 2013-10-21 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Syntaxin Binding Mechanism and Disease-Causing Mutations in Munc18-2
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2XHU
 
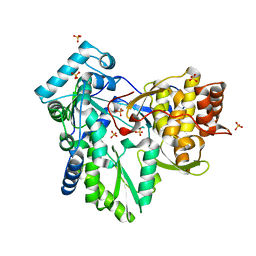 | | HCV-J4 NS5B Polymerase Orthorhombic Crystal Form | | Descriptor: | RNA-directed RNA polymerase, SULFATE ION | | Authors: | Harrus, D, Ahmed-El-Sayed, N, Simister, P.C, Miller, S, Triconnet, M, Hagedorn, C.H, Mahias, K, Rey, F.A, Astier-Gin, T, Bressanelli, S. | | Deposit date: | 2010-06-21 | | Release date: | 2010-08-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Further Insights Into the Roles of GTP and the C- Terminus of the Hepatitis C Virus Polymerase in the Initiation of RNA Synthesis
J.Biol.Chem., 285, 2010
|
|
2XI3
 
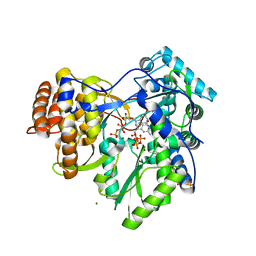 | | HCV-H77 NS5B Polymerase Complexed With GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNA-directed RNA polymerase | | Authors: | Harrus, D, Ahmed-El-Sayed, N, Simister, P.C, Miller, S, Triconnet, M, Hagedorn, C.H, Mahias, K, Rey, F.A, Astier-Gin, T, Bressanelli, S. | | Deposit date: | 2010-06-25 | | Release date: | 2010-08-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Further Insights Into the Roles of GTP and the C- Terminus of the Hepatitis C Virus Polymerase in the Initiation of RNA Synthesis
J.Biol.Chem., 285, 2010
|
|
2XI2
 
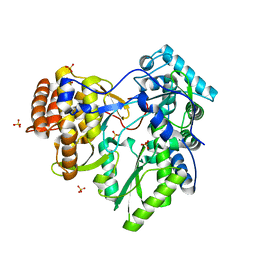 | | HCV-H77 NS5B Apo Polymerase | | Descriptor: | RNA-directed RNA polymerase, SULFATE ION | | Authors: | Harrus, D, Ahmed-El-Sayed, N, Simister, P.C, Miller, S, Triconnet, M, Hagedorn, C.H, Mahias, K, Rey, F.A, Astier-Gin, T, Bressanelli, S. | | Deposit date: | 2010-06-25 | | Release date: | 2010-08-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Further Insights Into the Roles of GTP and the C- Terminus of the Hepatitis C Virus Polymerase in the Initiation of RNA Synthesis
J.Biol.Chem., 285, 2010
|
|
2XHW
 
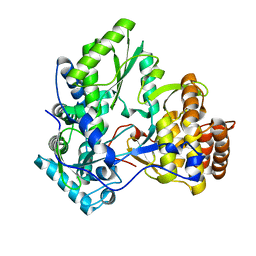 | | HCV-J4 NS5B Polymerase Trigonal Crystal Form | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Harrus, D, Ahmed-El-Sayed, N, Simister, P.C, Miller, S, Triconnet, M, Hagedorn, C.H, Mahias, K, Rey, F.A, Astier-Gin, T, Bressanelli, S. | | Deposit date: | 2010-06-21 | | Release date: | 2010-08-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Further Insights Into the Roles of GTP and the C- Terminus of the Hepatitis C Virus Polymerase in the Initiation of RNA Synthesis
J.Biol.Chem., 285, 2010
|
|
2XHV
 
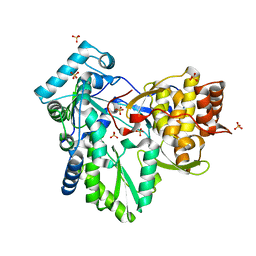 | | HCV-J4 NS5B Polymerase Point Mutant Orthorhombic Crystal Form | | Descriptor: | MAGNESIUM ION, RNA-directed RNA polymerase, SULFATE ION | | Authors: | Harrus, D, Ahmed-El-Sayed, N, Simister, P.C, Miller, S, Triconnet, M, Hagedorn, C.H, Mahias, K, Rey, F.A, Astier-Gin, T, Bressanelli, S. | | Deposit date: | 2010-06-21 | | Release date: | 2010-08-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Further Insights Into the Roles of GTP and the C- Terminus of the Hepatitis C Virus Polymerase in the Initiation of RNA Synthesis
J.Biol.Chem., 285, 2010
|
|
5LT9
 
 | | Ligand binding domain of Pseudomonas aeruginosa PAO1 amino acid chemoreceptors PctB in complex with L-Arg | | Descriptor: | ARGININE, GLYCEROL, Methyl-accepting chemotaxis protein PctB, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Ortega, A, Conejero-Muriel, M, Zhulin, I, Krell, T. | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
5LTO
 
 | | Ligand binding domain of Pseudomonas aeruginosa PAO1 amino acid chemoreceptors PctB in complex with L-Gln | | Descriptor: | GLUTAMINE, GLYCEROL, Methyl-accepting chemotaxis protein PctB, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Conejero-Muriel, M, Krell, T. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.459 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
5LTV
 
 | | LIGAND BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA PAO1 AMINO ACID CHEMORECPETOR PCTC IN COMPLEX WITH GABA | | Descriptor: | ACETATE ION, Chemotactic transducer PctC, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Conejero-Muriel, M, Krell, T. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
5LTX
 
 | | LIGAND BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA PAO1 AMINO ACID CHEMORECEPTOR PCTA IN COMPLEX WITH L-MET | | Descriptor: | ACETATE ION, Chemotaxis protein, FORMIC ACID, ... | | Authors: | Gavira, J.A, Rico-Gimenez, M, Ortega, A, Conejero-Muriel, M, Zhulin, I, Krell, T. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
5T7M
 
 | | LIGAND BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA PAO1 AMINO ACID CHEMORECEPTOR PCTA IN COMPLEX WITH L-TRP | | Descriptor: | ACETATE ION, Chemotaxis protein, SODIUM ION, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Ortega, A, Conejero-Muriel, M, Zhulin, I, Krell, T. | | Deposit date: | 2016-09-05 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
5T65
 
 | | LIGAND BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA PAO1 AMINO ACID CHEMORECEPTOR PCTA IN COMPLEX WITH L-ILE | | Descriptor: | ACETATE ION, ISOLEUCINE, Methyl-accepting chemotaxis protein PctA, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Ortega, A, Conejero-Muriel, M, Zhulin, I, Krell, T. | | Deposit date: | 2016-09-01 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
8QPD
 
 | | Structure of thioredoxin m from pea | | Descriptor: | Thioredoxin M-type, chloroplastic | | Authors: | Neira, J.L, Camara Artigas, A. | | Deposit date: | 2023-10-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure, dynamics and binding of thioredoxin m from Pisum sativum.
Int.J.Biol.Macromol., 262, 2024
|
|
2ERM
 
 | | Solution structure of a biologically active human FGF-1 monomer, complexed to a hexasaccharide heparin-analogue | | Descriptor: | 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, Heparin-binding growth factor 1, ISOPROPYL ALCOHOL | | Authors: | Canales, A, Lozano, R, Nieto, P.M, Martin-Lomas, M, Gimenez-Gallego, G, Jimenez-Barbero, J. | | Deposit date: | 2005-10-25 | | Release date: | 2006-10-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a human FGF-1 monomer, activated by a hexasaccharide heparin-analogue.
Febs J., 273, 2006
|
|
2KSS
 
 | | NMR structure of Myxococcus xanthus antirepressor CarS1 | | Descriptor: | Carotenogenesis protein carS | | Authors: | Jimenez, M, Gonzalez, C, Padmanabhan, S, Leon, E, Navarro-Aviles, G, Elias-Arnanz, M. | | Deposit date: | 2010-01-13 | | Release date: | 2010-05-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A bacterial antirepressor with SH3 domain topology mimics operator DNA in sequestering the repressor DNA recognition helix.
Nucleic Acids Res., 38, 2010
|
|
1HKN
 
 | | A complex between acidic fibroblast growth factor and 5-amino-2-naphthalenesulfonate | | Descriptor: | 5-AMINO-NAPHTALENE-2-MONOSULFONATE, HEPARIN-BINDING GROWTH FACTOR 1 | | Authors: | Fernandez-Tornero, C, Lozano, R.M, Gimenez-Gallego, G, Romero, A. | | Deposit date: | 2003-03-10 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Leads for Development of New Naphthalenesulfonate Derivatives with Enhanced Antiangiogenic Activity: Crystal Structure of Acidic Fibroblast Growth Factor in Complex with 5-Amino-2-Naphthalenesulfonate
J.Biol.Chem., 278, 2003
|
|
