4RLH
 
 | | Crystal structure of enoyl ACP reductase from Burkholderia pseudomallei in complex with AFN-1252 | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], N-methyl-N-[(3-methyl-1-benzofuran-2-yl)methyl]-3-(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-3-yl)propanamide | | Authors: | Rao, K.N, Sarah, J, Anirudha, L, Subramanya, H.S. | | Deposit date: | 2014-10-17 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of enoyl ACP reductase from
Burkholderia pseudomallei in complex with AFN-1252
To be Published
|
|
4O97
 
 | | Crystal structure of matriptase in complex with inhibitor | | Descriptor: | N-(trans-4-aminocyclohexyl)-3,5-bis[(3-carbamimidoylbenzyl)oxy]benzamide, Peptide CGLR, Suppressor of tumorigenicity 14 protein | | Authors: | Rao, K.N, Chandra, B.R, Ashok, K.N, Chakshusmathi, G, Ramesh, K.S, Subramanya, H.S. | | Deposit date: | 2014-01-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-guided discovery of 1,3,5 tri-substituted benzenes as potent and selective matriptase inhibitors exhibiting in vivo antitumor efficacy.
Bioorg.Med.Chem., 22, 2014
|
|
4O9V
 
 | | Crystal structure of matriptase in complex with inhibitor | | Descriptor: | N-(trans-4-aminocyclohexyl)-3,5-bis(4-carbamimidoylphenoxy)benzamide, Peptide CGLR, Suppressor of tumorigenicity 14 protein | | Authors: | Rao, K.N, Chandra, B.R, Ashok, K.N, Chakshusmathi, G, Ramesh, K.S, Subramanya, H.S. | | Deposit date: | 2014-01-03 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided discovery of 1,3,5 tri-substituted benzenes as potent and selective matriptase inhibitors exhibiting in vivo antitumor efficacy.
Bioorg.Med.Chem., 22, 2014
|
|
1U02
 
 | | Crystal structure of trehalose-6-phosphate phosphatase related protein | | Descriptor: | GLYCEROL, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Krishnamurthy, N.R, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-12 | | Release date: | 2004-07-20 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of trehalose-6-phosphate phosphatase-related protein: biochemical and biological implications.
Protein Sci., 15, 2006
|
|
1ZCC
 
 | | Crystal structure of glycerophosphodiester phosphodiesterase from Agrobacterium tumefaciens str.C58 | | Descriptor: | ACETATE ION, SULFATE ION, glycerophosphodiester phosphodiesterase | | Authors: | Krishnamurthy, N.R, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-04-11 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of glycerophosphodiester phosphodiesterase from Agrobacterium tumefaciens by SAD with a large asymmetric unit.
Proteins, 65, 2006
|
|
2M6F
 
 | | NMR solution structure of trans (major) form of In936 in Methanol | | Descriptor: | Contryphan-In | | Authors: | Sonti, R. | | Deposit date: | 2013-03-29 | | Release date: | 2013-10-30 | | Last modified: | 2022-08-24 | | Method: | SOLUTION NMR | | Cite: | Conformational diversity in contryphans from Conus venom: cis-trans isomerisation and aromatic/proline interactions in the 23-membered ring of a 7-residue peptide disulfide loop.
Chemistry, 19, 2013
|
|
2M6D
 
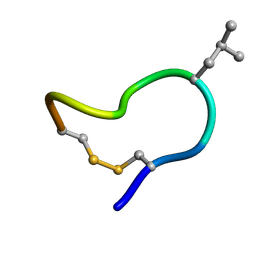 | | NMR solution structure of trans (major) form of In936 in water | | Descriptor: | Contryphan-In | | Authors: | Sonti, R. | | Deposit date: | 2013-03-28 | | Release date: | 2013-10-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conformational diversity in contryphans from Conus venom: cis-trans isomerisation and aromatic/proline interactions in the 23-membered ring of a 7-residue peptide disulfide loop.
Chemistry, 19, 2013
|
|
2M6C
 
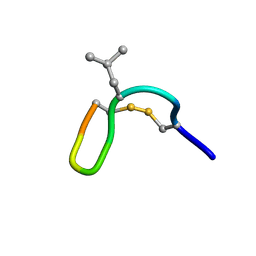 | | NMR solution structure of cis (minor) form of In936 in water | | Descriptor: | Contryphan-In | | Authors: | Sonti, R. | | Deposit date: | 2013-03-28 | | Release date: | 2013-10-30 | | Last modified: | 2022-08-24 | | Method: | SOLUTION NMR | | Cite: | Conformational diversity in contryphans from Conus venom: cis-trans isomerisation and aromatic/proline interactions in the 23-membered ring of a 7-residue peptide disulfide loop.
Chemistry, 19, 2013
|
|
2M6E
 
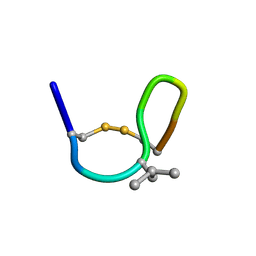 | | NMR solution structure of cis (minor) form of In936 in Methanol | | Descriptor: | Contryphan-In | | Authors: | Sonti, R. | | Deposit date: | 2013-03-29 | | Release date: | 2013-10-30 | | Last modified: | 2022-08-24 | | Method: | SOLUTION NMR | | Cite: | Conformational diversity in contryphans from Conus venom: cis-trans isomerisation and aromatic/proline interactions in the 23-membered ring of a 7-residue peptide disulfide loop.
Chemistry, 19, 2013
|
|
2M6H
 
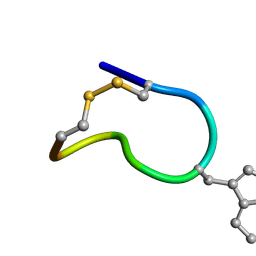 | | Solution structure of trans(C2-P3) trans (D5-P6) of LO959 in methanol | | Descriptor: | Contryphan-Lo | | Authors: | Sonti, R. | | Deposit date: | 2013-03-29 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Conformational diversity in contryphans from Conus venom: cis-trans isomerisation and aromatic/proline interactions in the 23-membered ring of a 7-residue peptide disulfide loop.
Chemistry, 19, 2013
|
|
2M6G
 
 | | Solution structure of cis(C2-P3) trans (D5-P6) form of lO959 in water | | Descriptor: | Contryphan-Lo | | Authors: | Sonti, R. | | Deposit date: | 2013-03-29 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Conformational diversity in contryphans from Conus venom: cis-trans isomerisation and aromatic/proline interactions in the 23-membered ring of a 7-residue peptide disulfide loop.
Chemistry, 19, 2013
|
|
4I8N
 
 | | CRYSTAL STRUCTURE of PROTEIN TYROSINE PHOSPHATASE 1B IN COMPLEX WITH AN INHIBITOR [(4-{(2S)-2-(1,3-BENZOXAZOL-2-YL)-2-[(4-FLUOROPHENYL)SULFAMOYL]ETHYL}PHENYL)AMINO](OXO)ACETIC ACID | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 1, [(4-{(2S)-2-(1,3-benzoxazol-2-yl)-2-[(4-fluorophenyl)sulfamoyl]ethyl}phenyl)amino](oxo)acetic acid | | Authors: | Reddy, S.M.V.V.V, Rao, K.N, Subramanya, H. | | Deposit date: | 2012-12-03 | | Release date: | 2012-12-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray Structure of PTP1B in Complex with a New PTP1B Inhibitor.
Protein Pept.Lett., 21, 2014
|
|
