2QUG
 
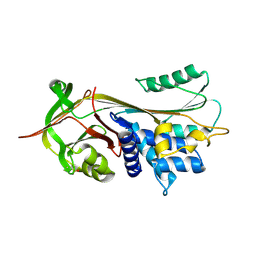 | | Crystal structure of alpha-1-antitrypsin, crystal form A | | Descriptor: | Alpha-1-antitrypsin | | Authors: | Hansen, G, Morton, C.J, Pearce, M.C, Feil, S.C, Adams, J.J, Parker, M.W, Bottomley, S.P. | | Deposit date: | 2007-08-05 | | Release date: | 2008-08-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Preventing serpin aggregation: The molecular mechanism of citrate action upon antitrypsin unfolding.
Protein Sci., 17, 2008
|
|
2R5G
 
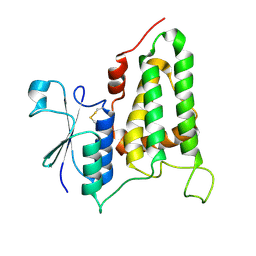 | | Structure of human CLIC2, crystal form B | | Descriptor: | Chloride intracellular channel protein 2 | | Authors: | Gorman, M.A, Hansen, G, Cromer, B.A, Parker, M.W. | | Deposit date: | 2007-09-03 | | Release date: | 2007-11-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of the Janus Protein Human CLIC2
J.Mol.Biol., 374, 2007
|
|
3L3V
 
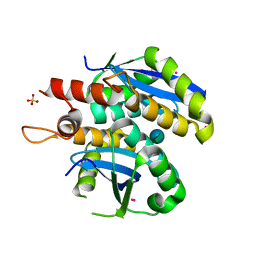 | | Structure of HIV-1 integrase core domain in complex with sucrose | | Descriptor: | CADMIUM ION, POL polyprotein, SULFATE ION, ... | | Authors: | Wielens, J, Chalmers, D.K, Scanlon, M.J, Parker, M.W. | | Deposit date: | 2009-12-18 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the HIV-1 integrase core domain in complex with sucrose reveals details of an allosteric inhibitory binding site
Febs Lett., 584, 2010
|
|
3L3U
 
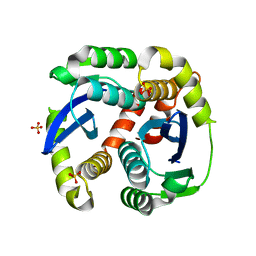 | | Crystal structure of the HIV-1 integrase core domain to 1.4A | | Descriptor: | POL polyprotein, SULFATE ION | | Authors: | Wielens, J, Chalmers, D.K, Scanlon, M.J, Parker, M.W. | | Deposit date: | 2009-12-17 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the HIV-1 integrase core domain in complex with sucrose reveals details of an allosteric inhibitory binding site.
Febs Lett., 584, 2010
|
|
1GSS
 
 | | THREE-DIMENSIONAL STRUCTURE OF CLASS PI GLUTATHIONE S-TRANSFERASE FROM HUMAN PLACENTA IN COMPLEX WITH S-HEXYLGLUTATHIONE AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | GLUTATHIONE S-TRANSFERASE, L-gamma-glutamyl-S-hexyl-L-cysteinylglycine | | Authors: | Reinemer, P, Dirr, H.W, Ladenstein, R, Lobello, M, Federici, G, Huber, R, Parker, M.W. | | Deposit date: | 1992-05-28 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of class pi glutathione S-transferase from human placenta in complex with S-hexylglutathione at 2.8 A resolution.
J.Mol.Biol., 227, 1992
|
|
4TKX
 
 | | Structure of Protease | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, LEAD (II) ION, ... | | Authors: | Gorman, M.A, Parker, M.W. | | Deposit date: | 2014-05-28 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the lysine specific protease Kgp from Porphyromonas gingivalis, a target for improved oral health.
Protein Sci., 24, 2015
|
|
4RS1
 
 | | Crystal structure of receptor-cytokine complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Granulocyte-macrophage colony-stimulating factor, ... | | Authors: | Broughton, S.E, Parker, M.W, Dhagat, U. | | Deposit date: | 2014-11-06 | | Release date: | 2015-11-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Conformational Changes in the GM-CSF Receptor Suggest a Molecular Mechanism for Affinity Conversion and Receptor Signaling.
Structure, 24, 2016
|
|
8G33
 
 | |
8G32
 
 | | Pro-form of a CDCL short from E. anophelis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Johnstone, B.A, Christie, M.P, Morton, C.J, Parker, M.W. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Pro-form of a CDCL short from E. anophelis
To Be Published
|
|
7TBA
 
 | | Pentraxin - ligand complex | | Descriptor: | C-reactive protein, CALCIUM ION, [3-(dibutylamino)propyl]phosphonic acid | | Authors: | Shing, K.S.C.T, Morton, C.J, Parker, M.W. | | Deposit date: | 2021-12-21 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A novel phosphocholine-mimetic inhibits a pro-inflammatory conformational change in C-reactive protein.
Embo Mol Med, 15, 2023
|
|
7L2Y
 
 | |
7L1P
 
 | | HIV Integrase Core domain (IN) in complex with dimer-spanning ligand | | Descriptor: | (2-{[3-(4-{2-[(3-{[3-(carboxymethyl)-5-methyl-1-benzofuran-2-yl]ethynyl}benzene-1-carbonyl)amino]ethyl}piperazine-1-carbonyl)phenyl]ethynyl}-5-methyl-1-benzofuran-3-yl)acetic acid, IODIDE ION, Integrase, ... | | Authors: | Gorman, M.A, Parker, M.W. | | Deposit date: | 2020-12-15 | | Release date: | 2021-12-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | HIV Integrase core domain in complex with inhibitor
To Be Published
|
|
1S3R
 
 | | Crystal structure of the human-specific toxin intermedilysin | | Descriptor: | SULFATE ION, intermedilysin | | Authors: | Polekhina, G, Giddings, K.S, Tweten, R.K, Parker, M.W. | | Deposit date: | 2004-01-14 | | Release date: | 2005-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into the action of the superfamily of cholesterol-dependent cytolysins from studies of intermedilysin
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2A2R
 
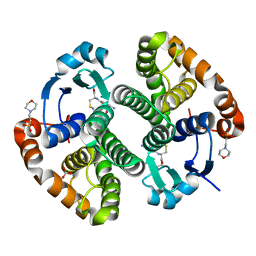 | | Crystal Structure of Glutathione Transferase Pi in complex with S-nitrosoglutathione | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-5-[1-(CARBOXYLATOMETHYLCARBAMOYL)-2-NITROSOSULFANYL-ETHYL]AMINO-5-OXO-PENTANOATE, CALCIUM ION, ... | | Authors: | Parker, L.J, Morton, C.J, Adams, J.J, Parker, M.W. | | Deposit date: | 2005-06-23 | | Release date: | 2006-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Calorimetric and structural studies of the nitric oxide carrier S-nitrosoglutathione bound to human glutathione transferase P1-1
Protein Sci., 15, 2006
|
|
1ZGN
 
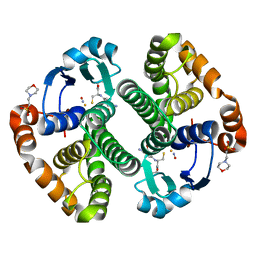 | | Crystal Structure of the Glutathione Transferase Pi in Complex with Dinitrosyl-diglutathionyl Iron Complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FE (III) ION, GLUTATHIONE, ... | | Authors: | Parker, L.J, Adams, J.J, Parker, M.W. | | Deposit date: | 2005-04-21 | | Release date: | 2005-11-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Nitrosylation of human glutathione transferase P1-1 with dinitrosyl diglutathionyl iron complex in vitro and in vivo
J.Biol.Chem., 280, 2005
|
|
2A2S
 
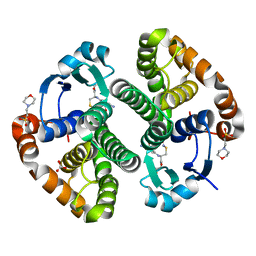 | | Crystal Structure of Human Glutathione Transferase in complex with S-nitrosoglutathione in the absence of reducing agent | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-5-[1-(CARBOXYLATOMETHYLCARBAMOYL)-2-NITROSOSULFANYL-ETHYL]AMINO-5-OXO-PENTANOATE, CALCIUM ION, ... | | Authors: | Parker, L.J, Morton, C.J, Adams, J.J, Parker, M.W. | | Deposit date: | 2005-06-23 | | Release date: | 2006-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Calorimetric and structural studies of the nitric oxide carrier S-nitrosoglutathione bound to human glutathione transferase P1-1
Protein Sci., 15, 2006
|
|
2GSS
 
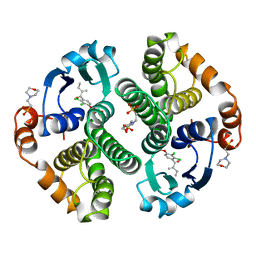 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1 IN COMPLEX WITH ETHACRYNIC ACID | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ETHACRYNIC ACID, GLUTATHIONE S-TRANSFERASE P1-1, ... | | Authors: | Oakley, A.J, Rossjohn, J, Parker, M.W. | | Deposit date: | 1996-10-29 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The three-dimensional structure of the human Pi class glutathione transferase P1-1 in complex with the inhibitor ethacrynic acid and its glutathione conjugate.
Biochemistry, 36, 1997
|
|
3N9J
 
 | |
6VQS
 
 | |
6VX2
 
 | |
6W0U
 
 | |
6W42
 
 | |
6PRO
 
 | | MnSOD from Geobacillus stearothermophilus | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Adams, J.J, Morton, C.J, Parker, M.W. | | Deposit date: | 2019-07-10 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.263 Å) | | Cite: | The Crystal Structure of the Manganese Superoxide Dismutase from Geobacillus stearothermophilus: Parker and Blake (1988) Revisited
Aust.J.Chem., 73, 2020
|
|
7RQ0
 
 | |
1Z0M
 
 | | the glycogen-binding domain of the AMP-activated protein kinase beta1 subunit | | Descriptor: | 5'-AMP-activated protein kinase, beta-1 subunit, Cycloheptakis-(1-4)-(alpha-D-glucopyranose) | | Authors: | Polekhina, G, Gupta, A, van Denderen, B.J, Feil, S.C, Kemp, B.E, Stapleton, D, Parker, M.W. | | Deposit date: | 2005-03-02 | | Release date: | 2005-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural Basis for Glycogen Recognition by AMP-Activated Protein Kinase.
Structure, 13, 2005
|
|
