3GTX
 
 | | D71G/E101G mutant in organophosphorus hydrolase from Deinococcus radiodurans | | Descriptor: | COBALT (II) ION, Organophosphorus hydrolase | | Authors: | Hawwa, R, Larsen, S, Ratia, K, Mesecar, A. | | Deposit date: | 2009-03-28 | | Release date: | 2009-06-30 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-based and random mutagenesis approaches increase the organophosphate-degrading activity of a phosphotriesterase homologue from Deinococcus radiodurans.
J.Mol.Biol., 393, 2009
|
|
3GTF
 
 | | D71G/E101G/V235L mutant in organophosphorus hydrolase from Deinococcus radiodurans | | Descriptor: | COBALT (II) ION, Organophosphorus hydrolase | | Authors: | Hawwa, R, Larsen, S, Ratia, K, Mesecar, A. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-30 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-based and random mutagenesis approaches increase the organophosphate-degrading activity of a phosphotriesterase homologue from Deinococcus radiodurans.
J.Mol.Biol., 393, 2009
|
|
3GU1
 
 | | Y97W mutant in organophosphorus hydrolase from Deinococcus radiodurans | | Descriptor: | COBALT (II) ION, GLYCEROL, Organophosphorus hydrolase | | Authors: | Hawwa, R, Larsen, S, Ratia, K, Mesecar, A. | | Deposit date: | 2009-03-28 | | Release date: | 2009-06-30 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based and random mutagenesis approaches increase the organophosphate-degrading activity of a phosphotriesterase homologue from Deinococcus radiodurans.
J.Mol.Biol., 393, 2009
|
|
3GU9
 
 | | R228A mutation in organophosphorus hydrolase from Deinococcus radiodurans | | Descriptor: | COBALT (II) ION, Organophosphorus hydrolase | | Authors: | Hawwa, R, Larsen, S, Ratia, K, Mesecar, A. | | Deposit date: | 2009-03-28 | | Release date: | 2009-06-30 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure-based and random mutagenesis approaches increase the organophosphate-degrading activity of a phosphotriesterase homologue from Deinococcus radiodurans.
J.Mol.Biol., 393, 2009
|
|
8SZ6
 
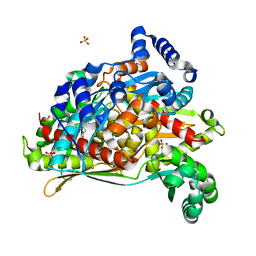 | | PmHMGR bound to mevaldehyde and CoA | | Descriptor: | (3R)-3,5,5-trihydroxy-3-methylpentanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, Mevaldyl-Coenzyme A, ... | | Authors: | Purohit, V, Stauffacher, C.V, Steussy, C.N. | | Deposit date: | 2023-05-27 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | pH-dependent reaction triggering in PmHMGR crystals for time-resolved crystallography.
Biophys.J., 123, 2024
|
|
3F4C
 
 | | Crystal structure of organophosphorus hydrolase from Geobacillus stearothermophilus strain 10, with glycerol bound | | Descriptor: | COBALT (II) ION, GLYCEROL, Organophosphorus hydrolase | | Authors: | Hawwa, R, Aikens, J, Turner, R.J, Santarsiero, B, Mesecar, A. | | Deposit date: | 2008-10-31 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis for thermostability revealed through the identification and characterization of a highly thermostable phosphotriesterase-like lactonase from Geobacillus stearothermophilus.
Arch.Biochem.Biophys., 488, 2009
|
|
3F4D
 
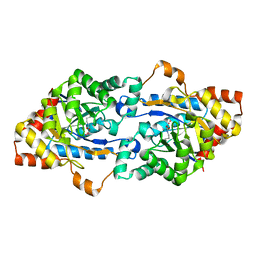 | | Crystal structure of organophosphorus hydrolase from Geobacillus stearothermophilus strain 10 | | Descriptor: | COBALT (II) ION, Organophosphorus hydrolase | | Authors: | Hawwa, R, Aikens, J, Turner, R.J, Santarsiero, B, Mesecar, A. | | Deposit date: | 2008-10-31 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural basis for thermostability revealed through the identification and characterization of a highly thermostable phosphotriesterase-like lactonase from Geobacillus stearothermophilus.
Arch.Biochem.Biophys., 488, 2009
|
|
8VLQ
 
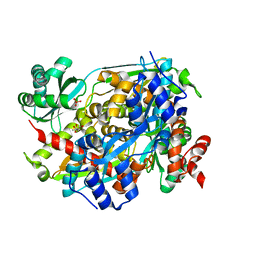 | | Structure of PmHMGR bound to mevalonate, CoA and NAD 5 minutes after reaction initiation at pH 9 | | Descriptor: | (R)-MEVALONATE, (R)-mevaldehyde, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Purohit, V, Steussy, C.N, Stauffacher, C.V. | | Deposit date: | 2024-01-12 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | pH-dependent reaction triggering in PmHMGR crystals for time-resolved crystallography.
Biophys.J., 123, 2024
|
|
7JRD
 
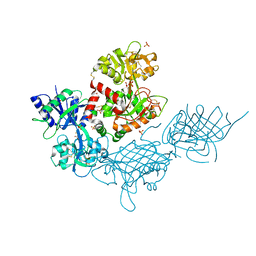 | |
7N88
 
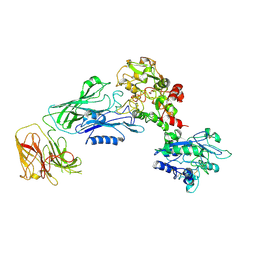 | |
6W02
 
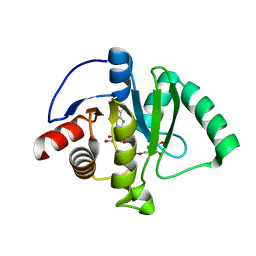 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in the complex with ADP ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | Authors: | Michalska, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-28 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
6VXS
 
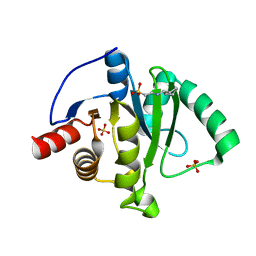 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Non-structural protein 3, ... | | Authors: | Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-24 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
6W6Y
 
 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in complex with AMP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, Non-structural protein 3 | | Authors: | Michalska, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-18 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
6WCF
 
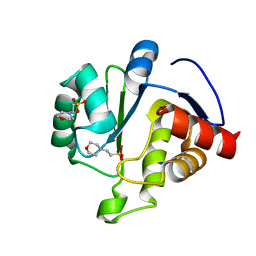 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS-CoV-2 in complex with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Non-structural protein 3 | | Authors: | Michalska, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-30 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.065 Å) | | Cite: | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
