1I8T
 
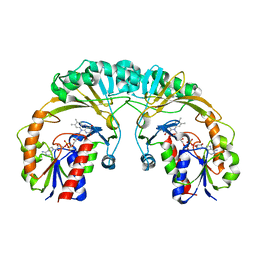 | | STRUCTURE OF UDP-GALACTOPYRANOSE MUTASE FROM E.COLI | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, UDP-GALACTOPYRANOSE MUTASE | | Authors: | Sanders, D.A.R, Staines, A.G, McMahon, S.A, McNeil, M.R, Whitfield, C, Naismith, J.H. | | Deposit date: | 2001-03-16 | | Release date: | 2001-10-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | UDP-galactopyranose mutase has a novel structure and mechanism.
Nat.Struct.Biol., 8, 2001
|
|
6ENK
 
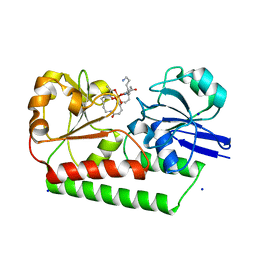 | | The X-ray crystal structure of DesE bound to desferrioxamine B | | Descriptor: | DesE, SODIUM ION, desferrioxamine B | | Authors: | Naismith, J.H, McMahon, S.A, Challis, G.L, Kadi, N, Oke, M, Liu, H, Carter, L.G, Johnson, K.A. | | Deposit date: | 2017-10-05 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Desferrioxamine biosynthesis: diverse hydroxamate assembly by substrate-tolerant acyl transferase DesC.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 373, 2018
|
|
6FI2
 
 | | VexL: A periplasmic depolymerase provides new insight into ABC transporter-dependent secretion of bacterial capsular polysaccharides | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranuronic acid-(1-4)-3-O-acetyl-2-acetamido-2-deoxy-alpha-D-galactopyranuronic acid-(1-4)-3-O-acetyl-2-acetamido-2-deoxy-alpha-D-galactopyranuronic acid, MALONATE ION, VexL | | Authors: | Naismith, J.H, McMahon, S.A, Le Bas, A, Liston, S.D, Whitfield, C. | | Deposit date: | 2018-01-16 | | Release date: | 2018-05-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Periplasmic depolymerase provides insight into ABC transporter-dependent secretion of bacterial capsular polysaccharides.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4FQ9
 
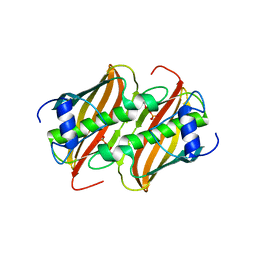 | | Crystal Structure of 3-hydroxydecanoyl-Acyl Carrier Protein Dehydratase (FabA) from Pseudomonas aeruginosa | | Descriptor: | 3-hydroxydecanoyl-[acyl-carrier-protein] dehydratase, GLYCEROL, PHOSPHATE ION | | Authors: | Moynie, L, Mcmahon, S.A, Duthie, F.G, Naismith, J.H. | | Deposit date: | 2012-06-25 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural insights into the mechanism and inhibition of the beta-hydroxydecanoyl-acyl carrier protein dehydratase from Pseudomonas aeruginosa
J.Mol.Biol., 425, 2013
|
|
5N4B
 
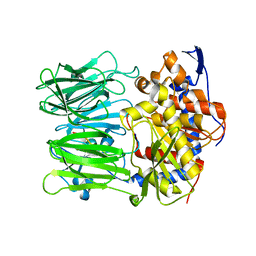 | | Prolyl oligopeptidase B from Galerina marginata bound to 25mer macrocyclization substrate - S577A mutant | | Descriptor: | Alpha-amanitin proprotein, Prolyl oligopeptidase | | Authors: | Czekster, C.M, McMahon, S.A, Ludewig, H, Naismith, J.H. | | Deposit date: | 2017-02-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Characterization of a dual function macrocyclase enables design and use of efficient macrocyclization substrates.
Nat Commun, 8, 2017
|
|
5N4E
 
 | | Prolyl oligopeptidase B from Galerina marginata bound to 35mer hydrolysis and macrocyclization substrate - H698A mutant | | Descriptor: | Alpha-amanitin proprotein, GLYCEROL, Prolyl oligopeptidase | | Authors: | Czekster, C.M, McMahon, S.A, Ludewig, H, Naismith, J.H. | | Deposit date: | 2017-02-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Characterization of a dual function macrocyclase enables design and use of efficient macrocyclization substrates.
Nat Commun, 8, 2017
|
|
5N4D
 
 | | Prolyl oligopeptidase B from Galerina marginata bound to 25mer macrocyclization substrate - D661A mutant | | Descriptor: | Alpha-amanitin proprotein, GLYCEROL, Prolyl oligopeptidase | | Authors: | Czekster, C.M, McMahon, S.A, Ludewig, H, Naismith, J.H. | | Deposit date: | 2017-02-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Characterization of a dual function macrocyclase enables design and use of efficient macrocyclization substrates.
Nat Commun, 8, 2017
|
|
5N4C
 
 | | Prolyl oligopeptidase B from Galerina marginata bound to 35mer hydrolysis and macrocyclization substrate - S577A mutant | | Descriptor: | Alpha-amanitin proprotein, GLYCEROL, Prolyl oligopeptidase | | Authors: | Czekster, C.M, McMahon, S.A, Ludewig, H, Naismith, J.H. | | Deposit date: | 2017-02-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Characterization of a dual function macrocyclase enables design and use of efficient macrocyclization substrates.
Nat Commun, 8, 2017
|
|
5N4F
 
 | | Prolyl oligopeptidase B from Galerina marginata - apo protein | | Descriptor: | GLYCEROL, Prolyl oligopeptidase | | Authors: | Czekster, C.M, McMahon, S.A, Ludewig, H, Naismith, J.H. | | Deposit date: | 2017-02-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization of a dual function macrocyclase enables design and use of efficient macrocyclization substrates.
Nat Commun, 8, 2017
|
|
3TEK
 
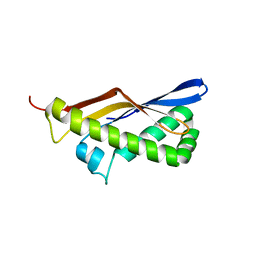 | | ThermoDBP: a non-canonical single-stranded DNA binding protein with a novel structure and mechanism | | Descriptor: | ThermoDBP-single stranded DNA binding protein | | Authors: | White, M.F, Paytubi, S, Liu, H, Graham, S, McMahon, S.A, Naismith, J.H. | | Deposit date: | 2011-08-15 | | Release date: | 2011-11-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Displacement of the canonical single-stranded DNA-binding protein in the Thermoproteales.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2XEU
 
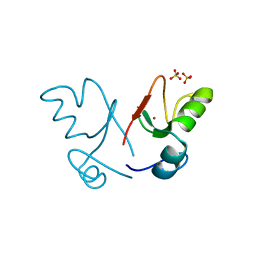 | | Ring domain | | Descriptor: | RING FINGER PROTEIN 4, SULFATE ION, ZINC ION, ... | | Authors: | Plechanovova, A, McMahon, S.A, Johnson, K.A, Navratilova, I, Naismith, J.H, Hay, R.T. | | Deposit date: | 2010-05-18 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanism of Ubiquitylation by Dimeric Ring Ligase Rnf4
Nat.Struct.Mol.Biol., 18, 2011
|
|
2XU2
 
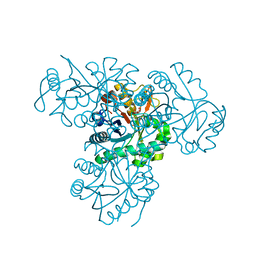 | | Crystal Structure of the hypothetical protein PA4511 from Pseudomonas aeruginosa | | Descriptor: | CITRIC ACID, UPF0271 PROTEIN PA4511 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, McMahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-10-14 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2XU8
 
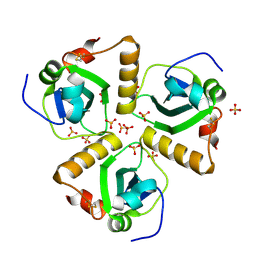 | | Structure of Pa1645 | | Descriptor: | PA1645, SULFATE ION | | Authors: | Abdelli, W.B, Moynie, L, McMahon, S.A, Liu, H, Alphey, M.S, Naismith, J.H. | | Deposit date: | 2010-10-15 | | Release date: | 2010-12-29 | | Last modified: | 2015-08-19 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery
Acta Crystallogr.,Sect.F, 69, 2013
|
|
2XBN
 
 | | Inhibition of the PLP-dependent enzyme serine palmitoyltransferase by cycloserine: evidence for a novel decarboxylative mechanism of inactivation | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, MAGNESIUM ION, SERINE PALMITOYLTRANSFERASE | | Authors: | Lowther, J, Yard, B.A, Johnson, K.A, Carter, L.G, Bhat, V.T, Raman, M.C.C, Clarke, D.J, Ramakers, B, McMahon, S.A, Naismith, J.H, Campopiano, D.J. | | Deposit date: | 2010-04-13 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibition of the Plp-Dependent Enzyme Serine Palmitoyltransferase by Cycloserine: Evidence for a Novel Decarboxylative Mechanism of Inactivation.
Mol.Biosystems, 6, 2010
|
|
7P1V
 
 | | Apo structure of KDNase from Trichophyton Rubrum | | Descriptor: | CALCIUM ION, Extracellular sialidase/neuraminidase, GLYCEROL | | Authors: | Gloster, T.M, McMahon, S.A. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Kinetic and Structural Characterization of Sialidases (Kdnases) from Ascomycete Fungal Pathogens.
Acs Chem.Biol., 16, 2021
|
|
7P1B
 
 | |
7P1E
 
 | | Structure of KDNase from Aspergillus Terrerus in complex with 2,3-difluoro-2-keto-3-deoxynononic acid | | Descriptor: | (2R,3R,4R,5R,6S)-2,3-bis(fluoranyl)-4,5-bis(oxidanyl)-6-[(1R,2R)-1,2,3-tris(oxidanyl)propyl]oxane-2-carboxylic acid, 3-deoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, CALCIUM ION, ... | | Authors: | Gloster, T.M, McMahon, S.A. | | Deposit date: | 2021-07-01 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Kinetic and Structural Characterization of Sialidases (Kdnases) from Ascomycete Fungal Pathogens.
Acs Chem.Biol., 16, 2021
|
|
7P1O
 
 | |
7P1S
 
 | | Structure of KDNase from Trichophyton Rubrum in complex with 2,3-didehydro-2,3-dideoxy-D-glycero-D-galacto-nonulosonic acid. | | Descriptor: | 2,6-anhydro-3-deoxy-D-glycero-D-galacto-non-2-enonic acid, Extracellular sialidase/neuraminidase, SODIUM ION | | Authors: | Gloster, T.M, McMahon, S.A. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Kinetic and Structural Characterization of Sialidases (Kdnases) from Ascomycete Fungal Pathogens.
Acs Chem.Biol., 16, 2021
|
|
7P1Q
 
 | |
7P1R
 
 | |
7P1D
 
 | |
7P1U
 
 | |
7P1F
 
 | | Structure of KDNase from Aspergillus terrerus in complex with 2,3-didehydro-2,3-dideoxy-D-glycero-D-galacto-nonulosonic acid. | | Descriptor: | 2,6-anhydro-3-deoxy-D-glycero-D-galacto-non-2-enonic acid, CALCIUM ION, GLYCEROL, ... | | Authors: | Gloster, T.M, McMahon, S.A. | | Deposit date: | 2021-07-01 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Kinetic and Structural Characterization of Sialidases (Kdnases) from Ascomycete Fungal Pathogens.
Acs Chem.Biol., 16, 2021
|
|
2IVY
 
 | | Crystal structure of hypothetical protein sso1404 from Sulfolobus solfataricus P2 | | Descriptor: | HYPOTHETICAL PROTEIN SSO1404 | | Authors: | Yan, X, Carter, L.G, Dorward, M, Liu, H, McMahon, S.A, Oke, M, Powers, H, White, M.F, Naismith, J.H. | | Deposit date: | 2006-06-22 | | Release date: | 2006-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
