1PWO
 
 | |
7YTB
 
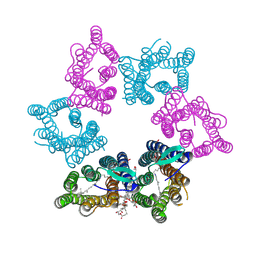 | | Crystal structure of Kin4B8 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Kin4B8, RETINAL | | Authors: | Murakoshi, S, Chazan, A, Shihoya, W, Beja, O, Nureki, O. | | Deposit date: | 2022-08-14 | | Release date: | 2023-03-15 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phototrophy by antenna-containing rhodopsin pumps in aquatic environments.
Nature, 615, 2023
|
|
6EEZ
 
 | |
4G8Y
 
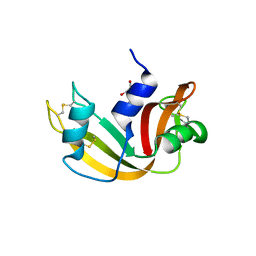 | | Crystal structure of Ribonuclease A in complex with 5b | | Descriptor: | 1-{[1-(alpha-L-arabinofuranosyl)-1H-1,2,3-triazol-4-yl]methyl}-5-methyl-2,4-dioxo-1,2,3,4-tetrahydropyrimidine, Ribonuclease pancreatic | | Authors: | Chatzileontiadou, D.S.M, Kantsadi, A.L, Leonidas, D.D. | | Deposit date: | 2012-07-23 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Triazole pyrimidine nucleosides as inhibitors of Ribonuclease A. Synthesis, biochemical, and structural evaluation.
Bioorg.Med.Chem., 20, 2012
|
|
4G90
 
 | | Crystal structure of Ribonuclease A in complex with 5e | | Descriptor: | 1-{[1-(alpha-L-arabinofuranosyl)-1H-1,2,3-triazol-4-yl]methyl}-5-fluoro-2,4-dioxo-1,2,3,4-tetrahydropyrimidine, Ribonuclease pancreatic | | Authors: | Chatzileontiadou, D.S.M, Kantsadi, A.L, Leonidas, D.D. | | Deposit date: | 2012-07-23 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Triazole pyrimidine nucleosides as inhibitors of Ribonuclease A. Synthesis, biochemical, and structural evaluation.
Bioorg.Med.Chem., 20, 2012
|
|
4G8V
 
 | | Crystal structure of Ribonuclease A in complex with 5a | | Descriptor: | 1-{[1-(alpha-L-arabinofuranosyl)-1H-1,2,3-triazol-4-yl]methyl}-2,4-dioxo-1,2,3,4-tetrahydropyrimidine, Ribonuclease pancreatic | | Authors: | Chatzileontiadou, D.S.M, Kantsadi, A.L, Leonidas, D.D. | | Deposit date: | 2012-07-23 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Triazole pyrimidine nucleosides as inhibitors of Ribonuclease A. Synthesis, biochemical, and structural evaluation.
Bioorg.Med.Chem., 20, 2012
|
|
1CA7
 
 | |
3T3H
 
 | | Glycogen Phosphorylase b in complex with GlcIU | | Descriptor: | 1-(beta-D-glucopyranosyl)-5-iodopyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2011-07-25 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The sigma-Hole Phenomenon of Halogen Atoms Forms the Structural Basis of the Strong Inhibitory Potency of C5 Halogen Substituted Glucopyranosyl Nucleosides towards Glycogen Phosphorylase b
Chemmedchem, 7, 2012
|
|
5LAR
 
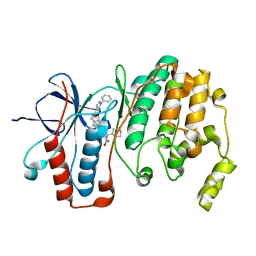 | | Crystal structure of p38 alpha MAPK14 in complex with VPC00628 | | Descriptor: | 5-azanyl-~{N}-[[4-[[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Petersen, L.K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel p38alpha MAP kinase inhibitors identified from yoctoReactor DNA-encoded small molecule library
Medchemcomm, 7, 2016
|
|
3T3I
 
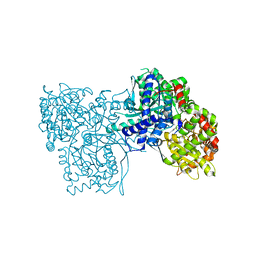 | | Glycogen Phosphorylase b in complex with GlcCF3U | | Descriptor: | 1-(beta-D-glucopyranosyl)-5-(trifluoromethyl)pyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2011-07-25 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The sigma-Hole Phenomenon of Halogen Atoms Forms the Structural Basis of the Strong Inhibitory Potency of C5 Halogen Substituted Glucopyranosyl Nucleosides towards Glycogen Phosphorylase b
Chemmedchem, 7, 2012
|
|
3SYR
 
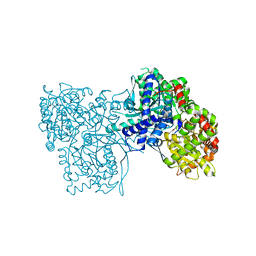 | | Glycogen phosphorylase b in complex with beta-D-glucopyranonucleoside 5-fluorouracil | | Descriptor: | 5-fluoro-1-(beta-D-glucopyranosyl)pyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Skamnaki, V.T, Kantsadi, A.L, Kontou, M, Leonidas, D.D. | | Deposit date: | 2011-07-18 | | Release date: | 2012-02-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3'-Axial CH(2) OH Substitution on Glucopyranose does not Increase Glycogen Phosphorylase Inhibitory Potency. QM/MM-PBSA Calculations Suggest Why.
Chem.Biol.Drug Des., 79, 2012
|
|
3T3D
 
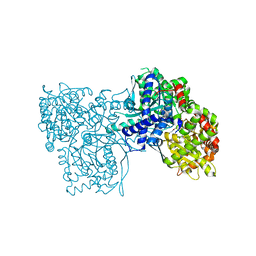 | | Glycogen phosphorylase b in complex with GlcU | | Descriptor: | 1-beta-D-glucopyranosylpyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2011-07-25 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The sigma-Hole Phenomenon of Halogen Atoms Forms the Structural Basis of the Strong Inhibitory Potency of C5 Halogen Substituted Glucopyranosyl Nucleosides towards Glycogen Phosphorylase b
Chemmedchem, 7, 2012
|
|
3SYM
 
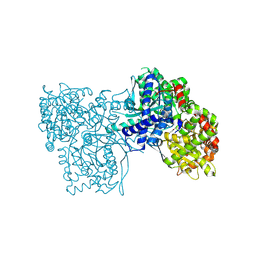 | | Glycogen Phosphorylase b in complex with 3 -C-(hydroxymethyl)-beta-D-glucopyranonucleoside of 5-fluorouracil | | Descriptor: | 5-fluoro-1-[3-C-(hydroxymethyl)-beta-D-glucopyranosyl]pyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Skamnaki, V.T, Katsandi, A.L, Kontou, M, Leonidas, D.D. | | Deposit date: | 2011-07-18 | | Release date: | 2012-02-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3'-Axial CH(2) OH Substitution on Glucopyranose does not Increase Glycogen Phosphorylase Inhibitory Potency. QM/MM-PBSA Calculations Suggest Why.
Chem.Biol.Drug Des., 79, 2012
|
|
3T3G
 
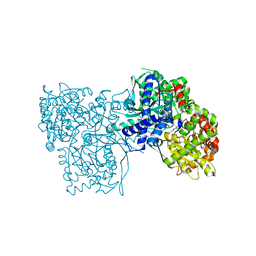 | | Glycogen Phosphorylase b in complex with GlcBrU | | Descriptor: | 5-bromo-1-(beta-D-glucopyranosyl)pyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2011-07-25 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The sigma-Hole Phenomenon of Halogen Atoms Forms the Structural Basis of the Strong Inhibitory Potency of C5 Halogen Substituted Glucopyranosyl Nucleosides towards Glycogen Phosphorylase b
Chemmedchem, 7, 2012
|
|
3T3E
 
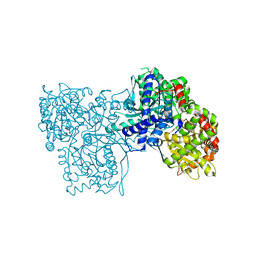 | | Glycogen phosphorylase b in complex with GlcClU | | Descriptor: | 5-chloro-1-(beta-D-glucopyranosyl)pyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2011-07-25 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The sigma-Hole Phenomenon of Halogen Atoms Forms the Structural Basis of the Strong Inhibitory Potency of C5 Halogen Substituted Glucopyranosyl Nucleosides towards Glycogen Phosphorylase b
Chemmedchem, 7, 2012
|
|
4ON1
 
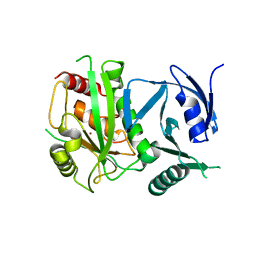 | | Crystal Structure of metalloproteinase-II from Bacteroides fragilis | | Descriptor: | GLYCEROL, Putative metalloprotease II, ZINC ION | | Authors: | Aleshin, A.E, Liddington, R.C, Shiryaev, S.A, Strongin, A.Y. | | Deposit date: | 2014-01-28 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural and functional diversity of metalloproteinases encoded by the Bacteroides fragilis pathogenicity island.
Febs J., 281, 2014
|
|
4EL5
 
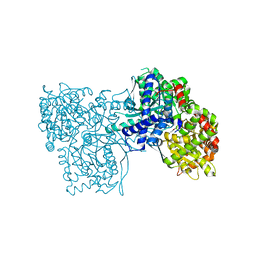 | | Crystal structure of GPb in complex with DK12 | | Descriptor: | 5-ethynyl-1-(beta-D-glucopyranosyl)pyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2012-04-10 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The binding of C5-alkynyl and alkylfurano[2,3-d]pyrimidine glucopyranonucleosides to glycogen phosphorylase b: Synthesis, biochemical and biological assessment.
Eur.J.Med.Chem., 54, 2012
|
|
4EJ2
 
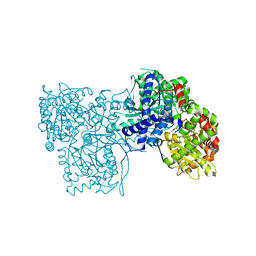 | | Crystal structure of GPb in complex with DK10 | | Descriptor: | 1-(beta-D-glucopyranosyl)-5-(hept-1-yn-1-yl)pyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2012-04-06 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The binding of C5-alkynyl and alkylfurano[2,3-d]pyrimidine glucopyranonucleosides to glycogen phosphorylase b: Synthesis, biochemical and biological assessment.
Eur.J.Med.Chem., 54, 2012
|
|
4EKE
 
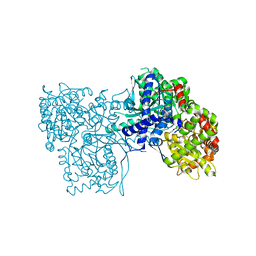 | | Crystal structure of GPb in complex with DK11 | | Descriptor: | 3-(beta-D-glucopyranosyl)-6-pentylfuro[2,3-d]pyrimidin-2(3H)-one, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2012-04-09 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The binding of C5-alkynyl and alkylfurano[2,3-d]pyrimidine glucopyranonucleosides to glycogen phosphorylase b: Synthesis, biochemical and biological assessment.
Eur.J.Med.Chem., 54, 2012
|
|
4EKY
 
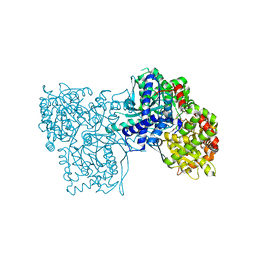 | | Crystal structure of GPb in complex with DK15 | | Descriptor: | 1-(beta-D-glucopyranosyl)-5-(pent-1-yn-1-yl)pyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2012-04-10 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The binding of C5-alkynyl and alkylfurano[2,3-d]pyrimidine glucopyranonucleosides to glycogen phosphorylase b: Synthesis, biochemical and biological assessment.
Eur.J.Med.Chem., 54, 2012
|
|
4EL0
 
 | | Crystal structure of GPb in complex with DK16 | | Descriptor: | 3-(beta-D-glucopyranosyl)-6-propylfuro[2,3-d]pyrimidin-2(3H)-one, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2012-04-10 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The binding of C5-alkynyl and alkylfurano[2,3-d]pyrimidine glucopyranonucleosides to glycogen phosphorylase b: Synthesis, biochemical and biological assessment.
Eur.J.Med.Chem., 54, 2012
|
|
1P1G
 
 | |
6WY1
 
 | | Crystal structure of an engineered thermostable dengue virus 2 envelope protein dimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dengue 2 soluble recombinant envelope | | Authors: | Kudlacek, S.T, Lakshmanane, P, Kuhlman, B. | | Deposit date: | 2020-05-12 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Designed, highly expressing, thermostable dengue virus 2 envelope protein dimers elicit quaternary epitope antibodies.
Sci Adv, 7, 2021
|
|
1CGQ
 
 | |
2VPL
 
 | | The structure of the complex between the first domain of L1 protein from Thermus thermophilus and mRNA from Methanococcus jannaschii | | Descriptor: | 50S RIBOSOMAL PROTEIN L1, FRAGMENT OF MRNA FOR L1-OPERON CONTAINING REGULATOR L1-BINDING SITE, POTASSIUM ION | | Authors: | Kljashtorny, V, Tishchenko, S, Nevskaya, N, Nikonov, S, Garber, M. | | Deposit date: | 2008-03-01 | | Release date: | 2008-09-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Domain II of Thermus thermophilus ribosomal protein L1 hinders recognition of its mRNA.
J. Mol. Biol., 383, 2008
|
|
