6XAS
 
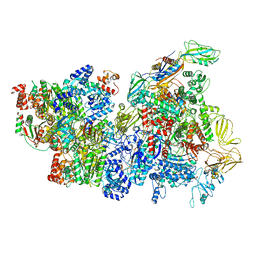 | | CryoEM Structure of E. coli Rho-dependent Transcription Pre-termination Complex | | Descriptor: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Hao, Z.T, Kim, H.K, Walz, T, Nudler, E. | | Deposit date: | 2020-06-04 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Pre-termination Transcription Complex: Structure and Function.
Mol.Cell, 81, 2021
|
|
6XAV
 
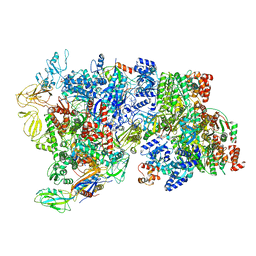 | | CryoEM Structure of E. coli Rho-dependent Transcription Pre-termination Complex bound with NusG | | Descriptor: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Hao, Z.T, Kim, H.K, Walz, T, Nudler, E. | | Deposit date: | 2020-06-04 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Pre-termination Transcription Complex: Structure and Function.
Mol.Cell, 81, 2021
|
|
1DGS
 
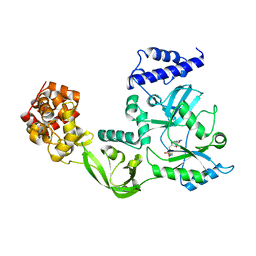 | | CRYSTAL STRUCTURE OF NAD+-DEPENDENT DNA LIGASE FROM T. FILIFORMIS | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA LIGASE, ZINC ION | | Authors: | Lee, J.Y, Chang, C, Song, H.K, Kwon, S.T, Suh, S.W. | | Deposit date: | 1999-11-25 | | Release date: | 2000-11-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications.
EMBO J., 19, 2000
|
|
2GJN
 
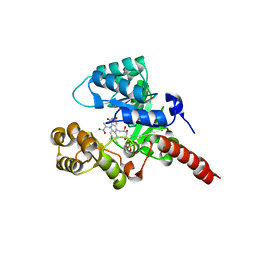 | |
2GJL
 
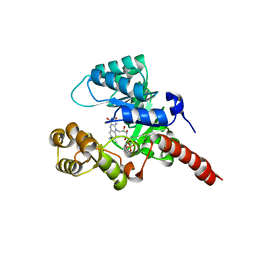 | | Crystal Structure of 2-nitropropane dioxygenase | | Descriptor: | FLAVIN MONONUCLEOTIDE, hypothetical protein PA1024 | | Authors: | Suh, S.W. | | Deposit date: | 2006-03-31 | | Release date: | 2006-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of 2-Nitropropane Dioxygenase Complexed with FMN and Substrate: identification of the catalytic base
J.Biol.Chem., 281, 2006
|
|
2ORY
 
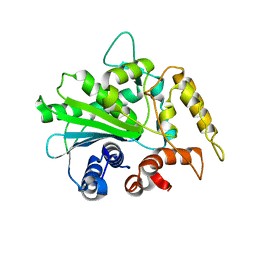 | | Crystal structure of M37 lipase | | Descriptor: | Lipase | | Authors: | Jung, S.K, Jeong, D.G, Kim, S.J. | | Deposit date: | 2007-02-05 | | Release date: | 2008-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the cold adaptation of psychrophilic M37 lipase from Photobacterium lipolyticum.
Proteins, 71, 2008
|
|
6JPV
 
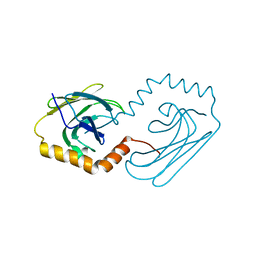 | | Structural analysis of AIMP2-DX2 and HSP70 interaction | | Descriptor: | Heat shock 70 kDa protein 1A,Aminoacyl tRNA synthase complex-interacting multifunctional protein 2 | | Authors: | Cho, H.Y, Son, S.Y, Jeon, Y.H. | | Deposit date: | 2019-03-28 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15000653 Å) | | Cite: | Targeting the interaction of AIMP2-DX2 with HSP70 suppresses cancer development.
Nat.Chem.Biol., 16, 2020
|
|
6K39
 
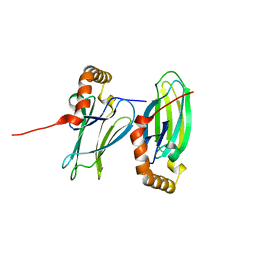 | | Structural analysis of AIMP2-DX2 and HSP70 interaction | | Descriptor: | Heat shock 70 kDa protein 1A,Aminoacyl tRNA synthase complex-interacting multifunctional protein 2 | | Authors: | Cho, H.Y, Son, S.Y, Jeon, Y.H. | | Deposit date: | 2019-05-16 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3981427 Å) | | Cite: | Targeting the interaction of AIMP2-DX2 with HSP70 suppresses cancer development.
Nat.Chem.Biol., 16, 2020
|
|
1KU0
 
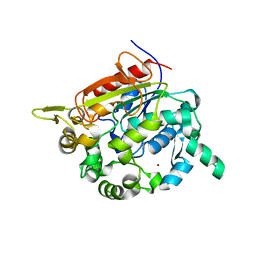 | | Structure of the Bacillus stearothermophilus L1 lipase | | Descriptor: | CALCIUM ION, L1 lipase, ZINC ION | | Authors: | Jeong, S.-T, Kim, H.-K, Kim, S.-J, Chi, S.-W, Pan, J.-G, Oh, T.-K, Ryu, S.-E. | | Deposit date: | 2002-01-18 | | Release date: | 2002-08-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel zinc-binding center and a temperature switch in the Bacillus stearothermophilus L1 lipase.
J.Biol.Chem., 277, 2002
|
|
7F3G
 
 | |
3A3W
 
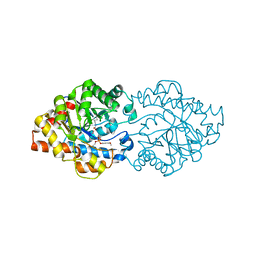 | | Structure of OpdA mutant (G60A/A80V/S92A/R118Q/K185R/Q206P/D208G/I260T/G273S) with diethyl 4-methoxyphenyl phosphate bound in the active site | | Descriptor: | COBALT (II) ION, DIETHYL 4-METHOXYPHENYL PHOSPHATE, Phosphotriesterase | | Authors: | Ollis, D.L, Tawfik, D.S, Schenk, G, Jackson, C.J, Foo, J.L, Tokuriki, N, Afriat, L, Carr, P.D, Kim, H.K. | | Deposit date: | 2009-06-23 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational sampling, catalysis, and evolution of the bacterial phosphotriesterase
Proc.Natl.Acad.Sci.USA, 2009
|
|
3A3X
 
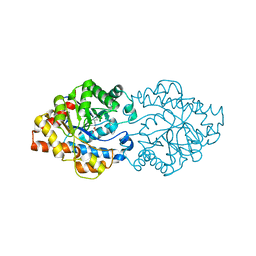 | | Structure of OpdA mutant (G60A/A80V/R118Q/K185R/Q206P/D208G/I260T/G273S) | | Descriptor: | COBALT (II) ION, Phosphotriesterase | | Authors: | Ollis, D.L, Tawfik, D.S, Schenk, G, Jackson, C.J, Foo, J.L, Tokuriki, N, Afriat, L, Carr, P.D, Kim, H.K. | | Deposit date: | 2009-06-23 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational sampling, catalysis, and evolution of the bacterial phosphotriesterase
Proc.Natl.Acad.Sci.USA, 2009
|
|
5XLN
 
 | |
1TA8
 
 | |
2DXL
 
 | |
2DXN
 
 | |
1G6Q
 
 | | CRYSTAL STRUCTURE OF YEAST ARGININE METHYLTRANSFERASE, HMT1 | | Descriptor: | HNRNP ARGININE N-METHYLTRANSFERASE | | Authors: | Weiss, V.H, McBride, A.E, Soriano, M.A, Filman, D.J, Silver, P.A, Hogle, J.M. | | Deposit date: | 2000-11-07 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure and oligomerization of the yeast arginine methyltransferase, Hmt1.
Nat.Struct.Biol., 7, 2000
|
|
