3BTS
 
 | |
4F3T
 
 | | Human Argonaute-2 - miR-20a complex | | Descriptor: | PHENOL, Protein argonaute-2, RNA (5'-R(P*UP*AP*AP*AP*GP*UP*GP*CP*UP*UP*AP*UP*AP*GP*UP*G*CP*AP*GP*G)-3') | | Authors: | Elkayam, E, Kuhn, C.-D, Tocilj, A, Joshua-Tor, L. | | Deposit date: | 2012-05-09 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Structure of Human Argonaute-2 in Complex with miR-20a.
Cell(Cambridge,Mass.), 150, 2012
|
|
3V2U
 
 | | Crystal structure of the yeast GAL regulon complex of the repressor, Gal80p, and the transducer, Gal3p, with galactose and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, Galactose/lactose metabolism regulatory protein GAL80, ... | | Authors: | Lavy, T, Kumar, P.R, He, H, Joshua-Tor, L. | | Deposit date: | 2011-12-12 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | The Gal3p transducer of the GAL regulon interacts with the Gal80p repressor in its ligand-induced closed conformation.
Genes Dev., 26, 2012
|
|
3BTU
 
 | |
4GGK
 
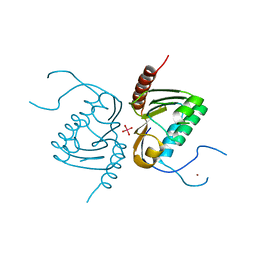 | | Crystal structure of Zucchini from mouse (mZuc / PLD6 / MitoPLD) bound to tungstate | | Descriptor: | Mitochondrial cardiolipin hydrolase, TUNGSTATE(VI)ION, ZINC ION | | Authors: | Ipsaro, J.J, Haase, A.D, Hannon, G.J, Joshua-Tor, L. | | Deposit date: | 2012-08-06 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis.
Nature, 491, 2012
|
|
3TIX
 
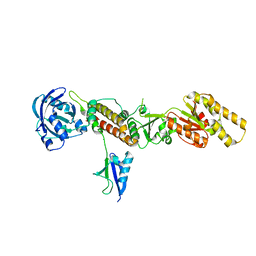 | | Crystal structure of the Chp1-Tas3 complex core | | Descriptor: | CHLORIDE ION, Chromo domain-containing protein 1, POTASSIUM ION, ... | | Authors: | Schalch, T, Joshua-Tor, L. | | Deposit date: | 2011-08-22 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9001 Å) | | Cite: | The Chp1-Tas3 core is a multifunctional platform critical for gene silencing by RITS.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3BTV
 
 | |
3V5R
 
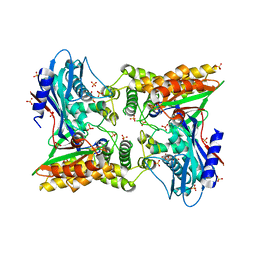 | | Crystal structure of the unliganded form of Gal3p | | Descriptor: | Protein GAL3, SULFATE ION | | Authors: | Lavy, T, Kumar, P.R, He, H, Joshua-Tor, L. | | Deposit date: | 2011-12-16 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | The Gal3p transducer of the GAL regulon interacts with the Gal80p repressor in its ligand-induced closed conformation.
Genes Dev., 26, 2012
|
|
4GGJ
 
 | | Crystal structure of Zucchini from mouse (mZuc / PLD6 / MitoPLD) | | Descriptor: | Mitochondrial cardiolipin hydrolase, ZINC ION | | Authors: | Ipsaro, J.J, Haase, A.D, Hannon, G.J, Joshua-Tor, L. | | Deposit date: | 2012-08-06 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis.
Nature, 491, 2012
|
|
1KQJ
 
 | | Crystal Structure of a Mutant of MutY Catalytic Domain | | Descriptor: | A/G-SPECIFIC ADENINE GLYCOSYLASE, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Messick, T.E, Chmiel, N.H, Golinelli, M.P, David, S.S, Joshua-Tor, L. | | Deposit date: | 2002-01-06 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Noncysteinyl coordination to the [4Fe-4S]2+ cluster of the DNA repair adenine glycosylase MutY introduced via site-directed mutagenesis. Structural characterization of an unusual histidinyl-coordinated cluster.
Biochemistry, 41, 2002
|
|
1KOU
 
 | | Crystal Structure of the Photoactive Yellow Protein Reconstituted with Caffeic Acid at 1.16 A Resolution | | Descriptor: | CAFFEIC ACID, N-BUTANE, PHOTOACTIVE YELLOW PROTEIN | | Authors: | van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 2001-12-22 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure of the photoactive yellow protein reconstituted with caffeic acid at 1.16 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6OU9
 
 | | Asymmetric focused reconstruction of human norovirus GI.7 Houston strain VLP asymmetric unit in T=3 symmetry | | Descriptor: | Major capsid protein | | Authors: | Jung, J, Grant, T, Thomas, D.R, Diehnelt, C.W, Grigorieff, N, Joshua-Tor, L. | | Deposit date: | 2019-05-04 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution cryo-EM structures of outbreak strain human norovirus shells reveal size variations.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OUT
 
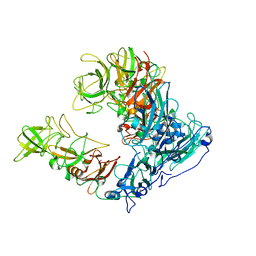 | | Asymmetric focused reconstruction of human norovirus GI.1 Norwalk strain VLP asymmetric unit in T=3 symmetry | | Descriptor: | Capsid protein VP1 | | Authors: | Jung, J, Grant, T, Thomas, D.R, Diehnelt, C.W, Grigorieff, N, Joshua-Tor, L. | | Deposit date: | 2019-05-05 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | High-resolution cryo-EM structures of outbreak strain human norovirus shells reveal size variations.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OTF
 
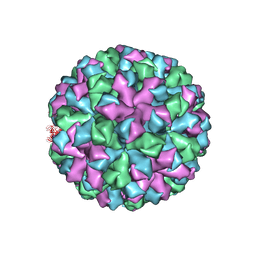 | | Symmetric reconstruction of human norovirus GII.2 Snow Mountain Virus Strain VLP in T=3 symmetry | | Descriptor: | Viral protein 1, ZINC ION | | Authors: | Jung, J, Grant, T, Thomas, D.R, Diehnelt, C.W, Grigorieff, N, Joshua-Tor, L. | | Deposit date: | 2019-05-03 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-resolution cryo-EM structures of outbreak strain human norovirus shells reveal size variations.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1KSY
 
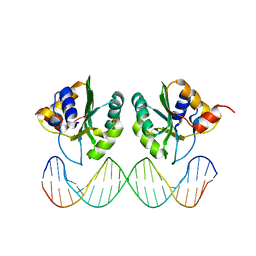 | | Crystal Structures of Two Intermediates in the Assembly of the Papillomavirus Replication Initiation Complex | | Descriptor: | E1 Recognition Sequence, Strand 1, Strand 2, ... | | Authors: | Enemark, E.J, Stenlund, A, Joshua-Tor, L. | | Deposit date: | 2002-01-14 | | Release date: | 2002-03-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structures of two intermediates in the assembly of the papillomavirus replication initiation complex.
EMBO J., 21, 2002
|
|
1KSX
 
 | |
6OUC
 
 | | Asymmetric focsued reconstruction of human norovirus GII.2 Snow Mountain Virus strain VLP asymmetric unit in T=1 symmetry | | Descriptor: | Viral protein 1, ZINC ION | | Authors: | Jung, J, Grant, T, Thomas, D.R, Diehnelt, C.W, Grigorieff, N, Joshua-Tor, L. | | Deposit date: | 2019-05-04 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | High-resolution cryo-EM structures of outbreak strain human norovirus shells reveal size variations.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OUU
 
 | | Symmetric reconstruction of human norovirus GII.4 Minerva strain VLP in T=4 symmetry | | Descriptor: | Major capsid protein | | Authors: | Jung, J, Grant, T, Thomas, D.R, Diehnelt, C.W, Grigorieff, N, Joshua-Tor, L. | | Deposit date: | 2019-05-05 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | High-resolution cryo-EM structures of outbreak strain human norovirus shells reveal size variations.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1CW9
 
 | | DNA DECAMER WITH AN ENGINEERED CROSSLINK IN THE MINOR GROOVE | | Descriptor: | 5'-D(*CP*CP*AP*GP*(G47)P*CP*CP*TP*GP*G)-3', CALCIUM ION | | Authors: | van Aalten, D.M.F, Erlanson, D.A, Verdine, G.L, Joshua-Tor, L. | | Deposit date: | 1999-08-26 | | Release date: | 1999-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A structural snapshot of base-pair opening in DNA.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1D7E
 
 | | CRYSTAL STRUCTURE OF THE P65 CRYSTAL FORM OF PHOTOACTIVE YELLOW PROTEIN | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 1999-10-17 | | Release date: | 2000-03-31 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Conformational substates in different crystal forms of the photoactive yellow protein--correlation with theoretical and experimental flexibility.
Protein Sci., 9, 2000
|
|
1CP2
 
 | | NITROGENASE IRON PROTEIN FROM CLOSTRIDIUM PASTEURIANUM | | Descriptor: | IRON/SULFUR CLUSTER, NITROGENASE IRON PROTEIN | | Authors: | Schlessman, J.L, Woo, D, Joshua-Tor, L, Howard, J.B, Rees, D.C. | | Deposit date: | 1998-05-11 | | Release date: | 1998-11-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Conformational variability in structures of the nitrogenase iron proteins from Azotobacter vinelandii and Clostridium pasteurianum.
J.Mol.Biol., 280, 1998
|
|
1GSV
 
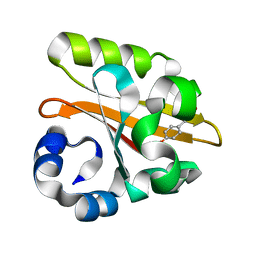 | | Crystal structure of the P65 crystal form of photoactive yellow protein G47S mutant | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 2002-01-08 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineering Photocycle Dynamics: Crystal Structures and Kinetics of Three Photoactive Yellow Protein Hinge-Bending Mutants
J.Biol.Chem., 227, 2002
|
|
1GSW
 
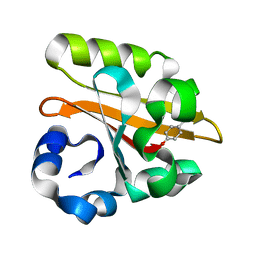 | | CRYSTAL STRUCTURE OF THE P65 CRYSTAL FORM OF PHOTOACTIVE YELLOW PROTEIN G51S MUTANT | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 2002-01-09 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering Photocycle Dynamics: Crystal Structures and Kinetics of Three Photoactive Yellow Protein Hinge-Bending Mutants
J.Biol.Chem., 227, 2002
|
|
1GSX
 
 | | CRYSTAL STRUCTURE OF THE P65 CRYSTAL FORM OF PHOTOACTIVE YELLOW PROTEIN G47S/G51S MUTANT | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 2002-01-09 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Engineering Photocycle Dynamics: Crystal Structures and Kinetics of Three Photoactive Yellow Protein Hinge-Bending Mutants
J.Biol.Chem., 227, 2002
|
|
1F08
 
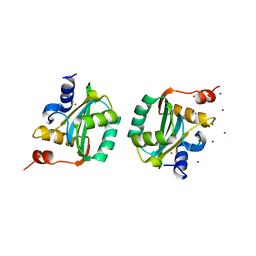 | | CRYSTAL STRUCTURE OF THE DNA-BINDING DOMAIN OF THE REPLICATION INITIATION PROTEIN E1 FROM PAPILLOMAVIRUS | | Descriptor: | BROMIDE ION, REPLICATION PROTEIN E1 | | Authors: | Enemark, E.J, Chen, G, Vaughn, D.E, Stenlund, A, Joshua-Tor, L. | | Deposit date: | 2000-05-15 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the DNA binding domain of the replication initiation protein E1 from papillomavirus.
Mol.Cell, 6, 2000
|
|
