2VBE
 
 | | Tailspike protein of bacteriophage Sf6 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Barbirz, S, Mueller, J.J, Freiberg, A, Heinemann, U, Seckler, R. | | Deposit date: | 2007-09-12 | | Release date: | 2008-04-22 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | An intersubunit active site between supercoiled parallel beta helices in the trimeric tailspike endorhamnosidase of Shigella flexneri Phage Sf6.
Structure, 16, 2008
|
|
2VJJ
 
 | | TAILSPIKE PROTEIN OF E.COLI BACTERIOPHAGE HK620 IN COMPLEX WITH HEXASACCHARIDE | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Mueller, J.J, Barbirz, S, Uetrecht, C, Seckler, R, Heinemann, U. | | Deposit date: | 2007-12-11 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal Structure of Escherichia Coli Phage Hk620 Tailspike: Podoviral Tailspike Endoglycosidase Modules are Evolutionarily Related.
Mol.Microbiol., 69, 2008
|
|
8AIF
 
 | |
5JDT
 
 | | Structure of Spin-labelled T4 lysozyme mutant L118C-R1 at 100K | | Descriptor: | AZIDE ION, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Loll, B, Consentius, P, Gohlke, U, Mueller, R, Kaupp, M, Heinemann, U, Wahl, M.C, Risse, T. | | Deposit date: | 2016-04-17 | | Release date: | 2016-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Tracking Transient Conformational States of T4 Lysozyme at Room Temperature Combining X-ray Crystallography and Site-Directed Spin Labeling.
J.Am.Chem.Soc., 138, 2016
|
|
5G27
 
 | | Structure of Spin-labelled T4 lysozyme mutant L118C-R1 at Room Temperature | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, ENDOLYSIN, ... | | Authors: | Gohlke, U, Consentius, P, Loll, B, Mueller, R, Kaupp, M, Heinemann, U, Risse, T. | | Deposit date: | 2016-04-07 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Tracking Transient Conformational States of T4 Lysozyme at Room Temperature Combining X-Ray Crystallography and Site-Directed Spin Labeling.
J.Am.Chem.Soc., 138, 2016
|
|
7ZIH
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor AG-01-128 | | Descriptor: | 8-(1~{H}-benzimidazol-2-ylmethyl)-3-ethyl-7-(phenylmethyl)purine-2,6-dione, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | Authors: | Schuetz, A, Gogolin, A, Pfeifer, J, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2022-04-08 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46890831 Å) | | Cite: | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7ZIK
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor LP533401 | | Descriptor: | (2~{R})-2-azanyl-3-[4-[2-azanyl-6-[(1~{R})-1-[4-chloranyl-2-(3-methylpyrazol-1-yl)phenyl]-2,2,2-tris(fluoranyl)ethoxy]pyrimidin-4-yl]phenyl]propanoic acid, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Schuetz, A, Heinemann, U. | | Deposit date: | 2022-04-08 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.58925915 Å) | | Cite: | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7ZII
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-05-193 | | Descriptor: | 8-(5~{H}-[1,3]dioxolo[4,5-f]benzimidazol-6-ylmethyl)-7-(phenylmethyl)-3-propyl-purine-2,6-dione, FE (III) ION, GLYCEROL, ... | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2022-04-08 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6280005 Å) | | Cite: | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7ZIJ
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-05-080 | | Descriptor: | 8-(1~{H}-benzimidazol-2-ylmethyl)-3-cyclopropyl-7-(phenylmethyl)purine-2,6-dione, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2022-04-08 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94678366 Å) | | Cite: | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7ZIG
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-05-060 | | Descriptor: | (2~{R})-2-azanyl-5-[[2-[[3-methyl-2,6-bis(oxidanylidene)-7-(phenylmethyl)purin-8-yl]methyl]-1~{H}-benzimidazol-5-yl]amino]-5-oxidanylidene-pentanoic acid, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2022-04-08 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.808885 Å) | | Cite: | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7ZIF
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-480 | | Descriptor: | (2R)-2-azanyl-5-[[2-[3-methyl-2,6-bis(oxidanylidene)-7-(phenylmethyl)purin-8-yl]sulfanyl-3H-benzimidazol-5-yl]amino]-5-oxidanylidene-pentanoic acid, FE (III) ION, GLYCEROL, ... | | Authors: | Schuetz, A, Ziebart, N, Weise, M, Mallow, K, Pfeifer, J, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2022-04-08 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.86859715 Å) | | Cite: | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
1MAC
 
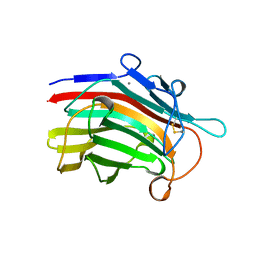 | | CRYSTAL STRUCTURE AND SITE-DIRECTED MUTAGENESIS OF BACILLUS MACERANS ENDO-1,3-1,4-BETA-GLUCANASE | | Descriptor: | 1,3-1,4-BETA-D-GLUCAN 4-GLUCANOHYDROLASE, CALCIUM ION | | Authors: | Hahn, M, Heinemann, U. | | Deposit date: | 1994-12-22 | | Release date: | 1995-02-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and site-directed mutagenesis of Bacillus macerans endo-1,3-1,4-beta-glucanase.
J.Biol.Chem., 270, 1995
|
|
6Q3V
 
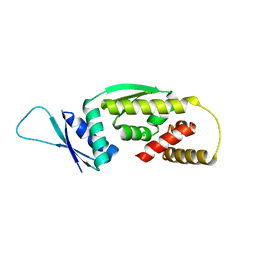 | |
2AYH
 
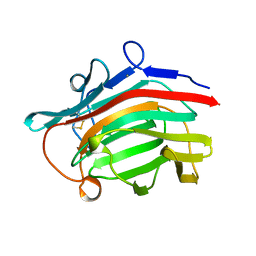 | | CRYSTAL AND MOLECULAR STRUCTURE AT 1.6 ANGSTROMS RESOLUTION OF THE HYBRID BACILLUS ENDO-1,3-1,4-BETA-D-GLUCAN 4-GLUCANOHYDROLASE H(A16-M) | | Descriptor: | 1,3-1,4-BETA-D-GLUCAN 4-GLUCANOHYDROLASE, CALCIUM ION | | Authors: | Hahn, M, Keitel, T, Heinemann, U. | | Deposit date: | 1995-02-02 | | Release date: | 1995-03-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal and molecular structure at 0.16-nm resolution of the hybrid Bacillus endo-1,3-1,4-beta-D-glucan 4-glucanohydrolase H(A16-M).
Eur.J.Biochem., 232, 1995
|
|
3FIL
 
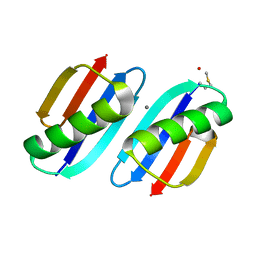 | |
3KXC
 
 | | Mutant transport protein | | Descriptor: | PALMITIC ACID, Trafficking protein particle complex subunit 3, Trafficking protein particle complex subunit 6B | | Authors: | Kummel, D, Heinemann, U. | | Deposit date: | 2009-12-02 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the self-palmitoylation activity of the transport protein particle component Bet3
Cell.Mol.Life Sci., 67, 2010
|
|
3JZ7
 
 | |
7OAS
 
 | | Structural basis for targeted p97 remodeling by ASPL as prerequisite for p97 trimethylation by METTL21D | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, S-adenosyl-L-methionine-dependent methyltransferases superfamily protein | | Authors: | Petrovic, S, Heinemann, U, Roske, Y. | | Deposit date: | 2021-04-20 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | Structural remodeling of AAA+ ATPase p97 by adaptor protein ASPL facilitates posttranslational methylation by METTL21D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7OAT
 
 | | Structural basis for targeted p97 remodelling by ASPL as prerequisite for p97 trimethylation by METTL21D | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Petrovic, S, Heinemann, U, Roske, Y. | | Deposit date: | 2021-04-20 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.995 Å) | | Cite: | Structural remodeling of AAA+ ATPase p97 by adaptor protein ASPL facilitates posttranslational methylation by METTL21D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1CSQ
 
 | |
1CPN
 
 | |
1CSP
 
 | |
1CPM
 
 | |
6TGF
 
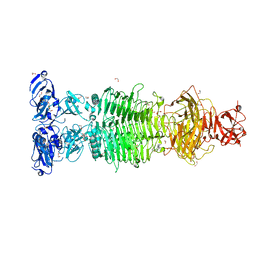 | | Pantoea stewartii WceF is a glycan biofilm modifying enzyme with a bacteriophage tailspike-like parallel beta-helix fold | | Descriptor: | 1,2-ETHANEDIOL, Exopolysaccharide biosynthesis protein, TETRAETHYLENE GLYCOL | | Authors: | Irmscher, T, Roske, Y, Gayk, I, Heinemann, U, Barbirz, S. | | Deposit date: | 2019-11-15 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Pantoea stewartii WceF is a glycan biofilm-modifying enzyme with a bacteriophage tailspike-like fold.
J.Biol.Chem., 296, 2021
|
|
1MJC
 
 | |
