3ZW6
 
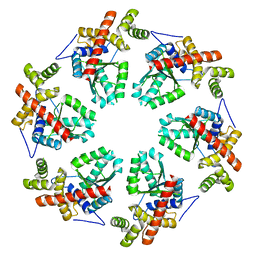 | | MODEL OF HEXAMERIC AAA DOMAIN ARRANGEMENT OF GREEN-TYPE RUBISCO ACTIVASE FROM TOBACCO. | | Descriptor: | RIBULOSE BISPHOSPHATE CARBOXYLASE/OXYGENASE ACTIVASE 1, CHLOROPLASTIC | | Authors: | Stotz, M, Mueller-Cajar, O, Ciniawsky, S, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2011-07-28 | | Release date: | 2011-11-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Structure of Green-Type Rubisco Activase from Tobacco
Nat.Struct.Mol.Biol., 18, 2011
|
|
3ZUH
 
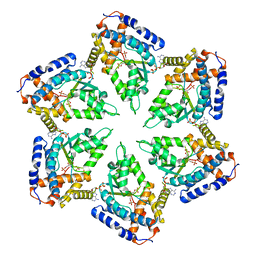 | | Negative stain EM Map of the AAA protein CbbX, a red-type Rubisco activase from R. sphaeroides | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PROTEIN CBBX, RIBULOSE-1,5-DIPHOSPHATE | | Authors: | Mueller-Cajar, O, Stotz, M, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2011-07-19 | | Release date: | 2011-11-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (21 Å) | | Cite: | Structure and Function of the Aaa+ Protein Cbbx, a Red-Type Rubisco Activase.
Nature, 479, 2011
|
|
6EKC
 
 | | Crystal structure of the BSD2 homolog of Arabidopsis thaliana bound to the octameric assembly of RbcL from Thermosynechococcus elongatus | | Descriptor: | DnaJ/Hsp40 cysteine-rich domain superfamily protein, Ribulose bisphosphate carboxylase large chain, ZINC ION | | Authors: | Aigner, H, Wilson, R.H, Bracher, A, Calisse, L, Bhat, J.Y, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2017-09-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Plant RuBisCo assembly in E. coli with five chloroplast chaperones including BSD2.
Science, 358, 2017
|
|
6EKB
 
 | | Crystal structure of the BSD2 homolog of Arabidopsis thaliana | | Descriptor: | DnaJ/Hsp40 cysteine-rich domain superfamily protein, ZINC ION | | Authors: | Aigner, H, Wilson, R.H, Bracher, A, Calisse, L, Bhat, J.Y, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2017-09-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Plant RuBisCo assembly in E. coli with five chloroplast chaperones including BSD2.
Science, 358, 2017
|
|
6Z1F
 
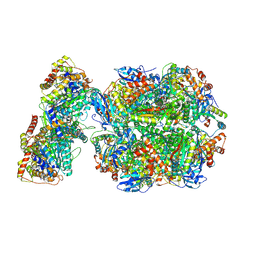 | | CryoEM structure of Rubisco Activase with its substrate Rubisco from Nostoc sp. (strain PCC7120) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wang, H, Bracher, A, Flecken, M, Popilka, L, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2020-05-13 | | Release date: | 2020-09-23 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Dual Functions of a Rubisco Activase in Metabolic Repair and Recruitment to Carboxysomes.
Cell, 183, 2020
|
|
6Z1G
 
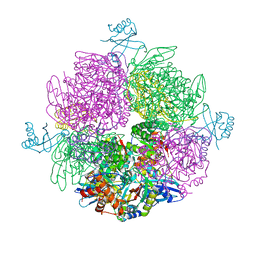 | | CryoEM structure of the interaction between Rubisco Activase small-subunit-like (SSUL) domain with Rubisco from Nostoc sp. (strain PCC7120) | | Descriptor: | Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain, Ribulose bisphosphate carboxylase/oxygenase activase | | Authors: | Wang, H, Bracher, A, Flecken, M, Popilka, L, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2020-05-13 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Dual Functions of a Rubisco Activase in Metabolic Repair and Recruitment to Carboxysomes.
Cell, 183, 2020
|
|
2PEN
 
 | | Crystal structure of RbcX, crystal form I | | Descriptor: | ORF134 | | Authors: | Saschenbrecker, S, Bracher, A, Vasudeva Rao, K, Vasudeva Rao, B, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2007-04-03 | | Release date: | 2007-07-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and Function of RbcX, an Assembly Chaperone for Hexadecameric Rubisco.
Cell(Cambridge,Mass.), 129, 2007
|
|
2PEJ
 
 | | Crystal structure of RbcX point mutant Y17A/Y20L | | Descriptor: | ORF134 | | Authors: | Saschenbrecker, S, Bracher, A, Vasudeva Rao, K, Vasudeva Rao, B, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2007-04-03 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and Function of RbcX, an Assembly Chaperone for Hexadecameric Rubisco.
Cell(Cambridge,Mass.), 129, 2007
|
|
2PEO
 
 | | Crystal structure of RbcX from Anabaena CA | | Descriptor: | RbcX protein | | Authors: | Saschenbrecker, S, Bracher, A, Vasudeva Rao, K, Vasudeva Rao, B, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2007-04-03 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Function of RbcX, an Assembly Chaperone for Hexadecameric Rubisco.
Cell(Cambridge,Mass.), 129, 2007
|
|
2PEK
 
 | | Crystal structure of RbcX point mutant Q29A | | Descriptor: | ORF134 | | Authors: | Saschenbrecker, S, Bracher, A, Vasudeva Rao, K, Vasudeva Rao, B, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2007-04-03 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure and Function of RbcX, an Assembly Chaperone for Hexadecameric Rubisco.
Cell(Cambridge,Mass.), 129, 2007
|
|
2PEM
 
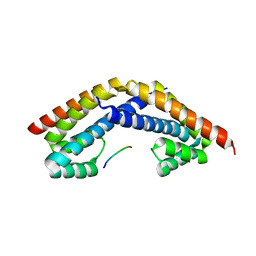 | | Crystal structure of RbcX in complex with substrate | | Descriptor: | ORF134, RbcL | | Authors: | Saschenbrecker, S, Bracher, A, Vasudeva Rao, K, Vasudeva Rao, B, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2007-04-03 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure and Function of RbcX, an Assembly Chaperone for Hexadecameric Rubisco.
Cell(Cambridge,Mass.), 129, 2007
|
|
2PEQ
 
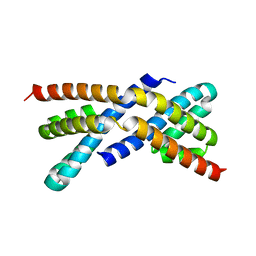 | | Crystal structure of RbcX, crystal form II | | Descriptor: | ORF134 | | Authors: | Saschenbrecker, S, Bracher, A, Vasudeva Rao, K, Vasudeva Rao, B, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2007-04-03 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Function of RbcX, an Assembly Chaperone for Hexadecameric Rubisco.
Cell(Cambridge,Mass.), 129, 2007
|
|
2PEI
 
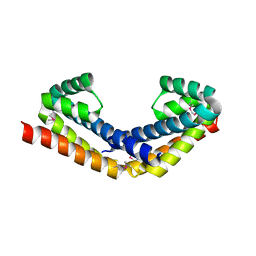 | | Crystal structure of selenomethionine-labeled RbcX | | Descriptor: | ORF134 | | Authors: | Saschenbrecker, S, Bracher, A, Vasudeva Rao, K, Vasudeva Rao, B, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2007-04-03 | | Release date: | 2007-07-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and Function of RbcX, an Assembly Chaperone for Hexadecameric Rubisco.
Cell(Cambridge,Mass.), 129, 2007
|
|
1ELR
 
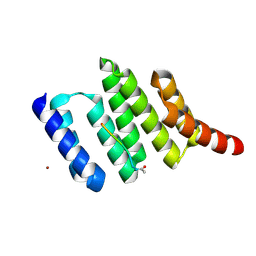 | | Crystal structure of the TPR2A domain of HOP in complex with the HSP90 peptide MEEVD | | Descriptor: | HSP90-PEPTIDE MEEVD, NICKEL (II) ION, TPR2A-DOMAIN OF HOP | | Authors: | Scheufler, C, Brinker, A, Hartl, F.U, Moarefi, I. | | Deposit date: | 2000-03-14 | | Release date: | 2000-04-26 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine.
Cell(Cambridge,Mass.), 101, 2000
|
|
1ELW
 
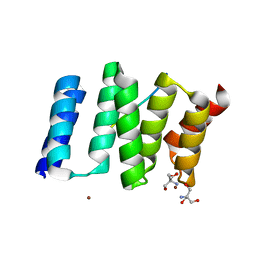 | | Crystal structure of the TPR1 domain of HOP in complex with a HSC70 peptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HSC70-PEPTIDE, NICKEL (II) ION, ... | | Authors: | Scheufler, C, Brinker, A, Hartl, F.U, Moarefi, I. | | Deposit date: | 2000-03-14 | | Release date: | 2000-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine
Cell(Cambridge,Mass.), 101, 2000
|
|
1XQS
 
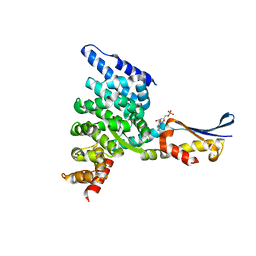 | | Crystal structure of the HspBP1 core domain complexed with the fragment of Hsp70 ATPase domain | | Descriptor: | ADENOSINE MONOPHOSPHATE, HSPBP1 protein, Heat shock 70 kDa protein 1 | | Authors: | Shomura, Y, Dragovic, Z, Chang, H.C, Tzvetkov, N, Young, J.C, Brodsky, J.L, Guerriero, V, Hartl, F.U, Bracher, A. | | Deposit date: | 2004-10-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Regulation of Hsp70 Function by HspBP1; Structural Analysis Reveals an Alternate Mechanism for Hsp70 Nucleotide Exchange
Mol.Cell, 17, 2005
|
|
1XQR
 
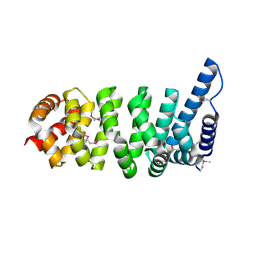 | | Crystal structure of the HspBP1 core domain | | Descriptor: | HspBP1 protein | | Authors: | Shomura, Y, Dragovic, Z, Chang, H.C, Tzvetkov, N, Young, J.C, Brodsky, J.L, Guerriero, V, Hartl, F.U, Bracher, A. | | Deposit date: | 2004-10-13 | | Release date: | 2005-03-01 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Regulation of Hsp70 Function by HspBP1; Structural Analysis Reveals an Alternate Mechanism for Hsp70 Nucleotide Exchange
Mol.Cell, 17, 2005
|
|
4V94
 
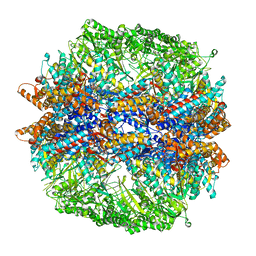 | | Molecular architecture of the eukaryotic chaperonin TRiC/CCT derived by a combination of chemical crosslinking and mass-spectrometry, XL-MS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Leitner, A, Joachimiak, L.A, Bracher, A, Walzthoeni, T, Chen, B, Monkemeyer, L, Pechmann, S, Holmes, S, Cong, Y, Ma, B, Ludtke, S, Chiu, W, Hartl, F.U, Aebersold, R, Frydman, J. | | Deposit date: | 2012-01-11 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Molecular Architecture of the Eukaryotic Chaperonin TRiC/CCT.
Structure, 20, 2012
|
|
6HBA
 
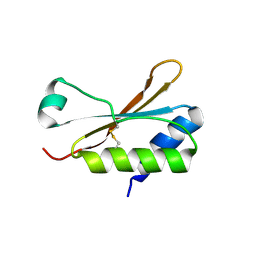 | | Crystal Structure of the small subunit-like domain 1 of CcmM from Synechococcus elongatus (strain PCC 7942), thiol-oxidized form | | Descriptor: | Carbon dioxide concentrating mechanism protein CcmM | | Authors: | Wang, H, Yan, X, Aigner, H, Bracher, A, Nguyen, N.D, Hee, W.Y, Long, B.M, Price, G.D, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Rubisco condensate formation by CcmM in beta-carboxysome biogenesis.
Nature, 566, 2019
|
|
6HBC
 
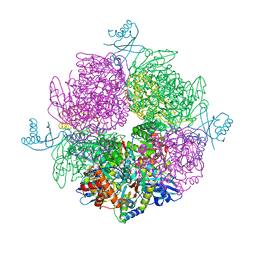 | | Structure of the repeat unit in the network formed by CcmM and Rubisco from Synechococcus elongatus | | Descriptor: | Carbon dioxide concentrating mechanism protein CcmM, Ribulose 1,5-bisphosphate carboxylase small subunit, Ribulose bisphosphate carboxylase large chain | | Authors: | Wang, H, Yan, X, Aigner, H, Bracher, A, Nguyen, N.D, Hee, W.Y, Long, B.M, Price, G.D, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Rubisco condensate formation by CcmM in beta-carboxysome biogenesis.
Nature, 566, 2019
|
|
6HBB
 
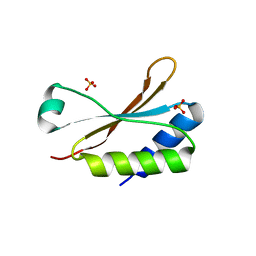 | | Crystal Structure of the small subunit-like domain 1 of CcmM from Synechococcus elongatus (strain PCC 7942) | | Descriptor: | Carbon dioxide concentrating mechanism protein CcmM, SULFATE ION | | Authors: | Wang, H, Yan, X, Aigner, H, Bracher, A, Nguyen, N.D, Hee, W.Y, Long, B.M, Price, G.D, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Rubisco condensate formation by CcmM in beta-carboxysome biogenesis.
Nature, 566, 2019
|
|
6EPC
 
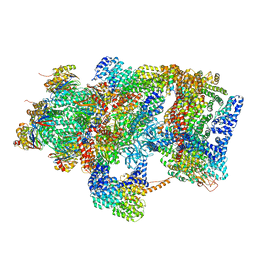 | | Ground state 26S proteasome (GS2) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (12.3 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
6EPD
 
 | | Substrate processing state 26S proteasome (SPS1) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (15.4 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
6EPF
 
 | | Ground state 26S proteasome (GS1) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (11.8 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
6EPE
 
 | | Substrate processing state 26S proteasome (SPS2) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (12.8 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
